05.05.2025 | Biodiversity and People, Media Release
Citizen Science project FLOW receives new funding
More30.04.2025 | Biodiversity Conservation, Media Release
From the front garden to the continent: Why biodiversity does not increase evenly from small to large
More29.04.2025 | iDiv, iDiv Members
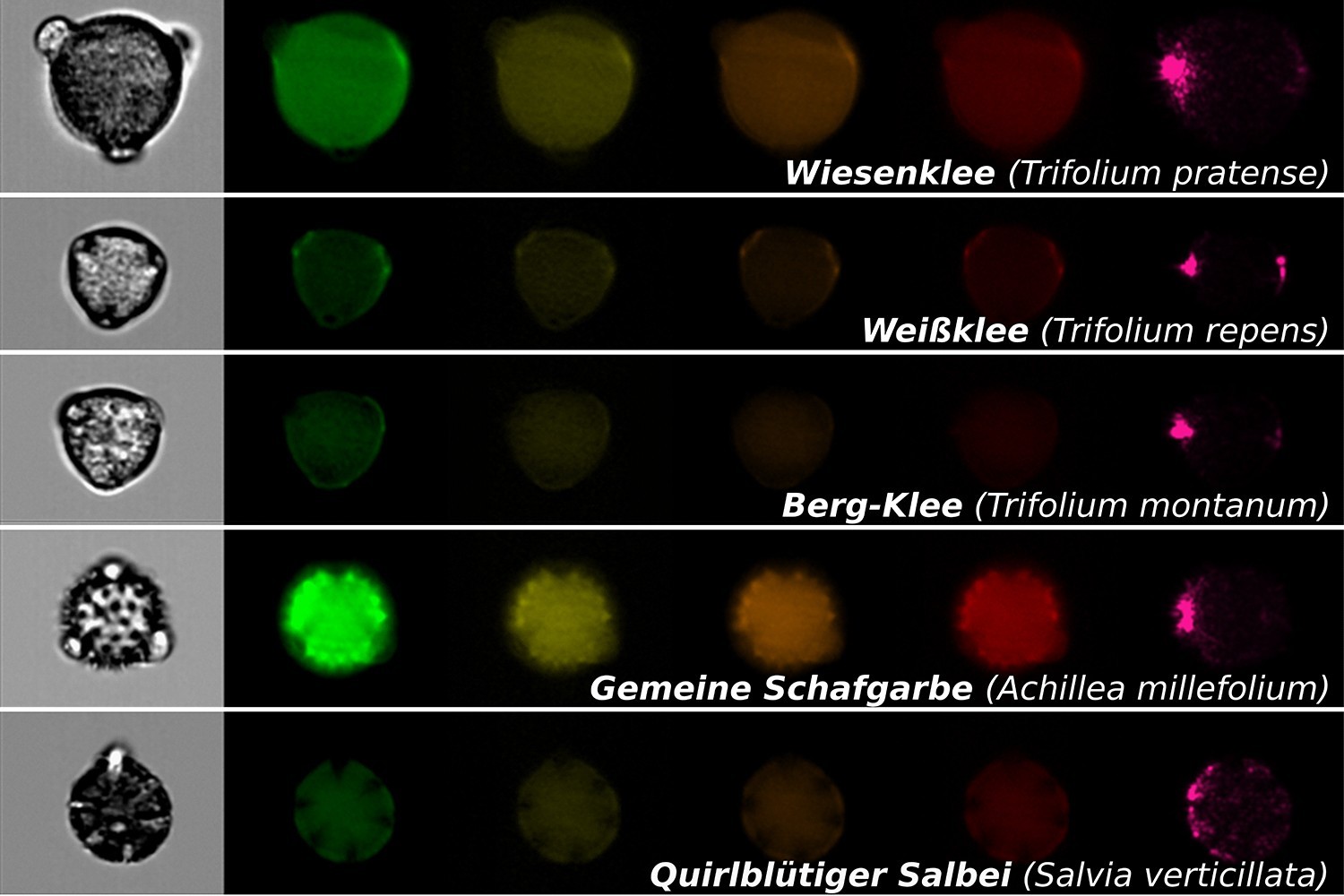
What microorganisms in tree crowns reveal about forest biodiversity
More17.04.2025 | iDiv, Media Release, Research
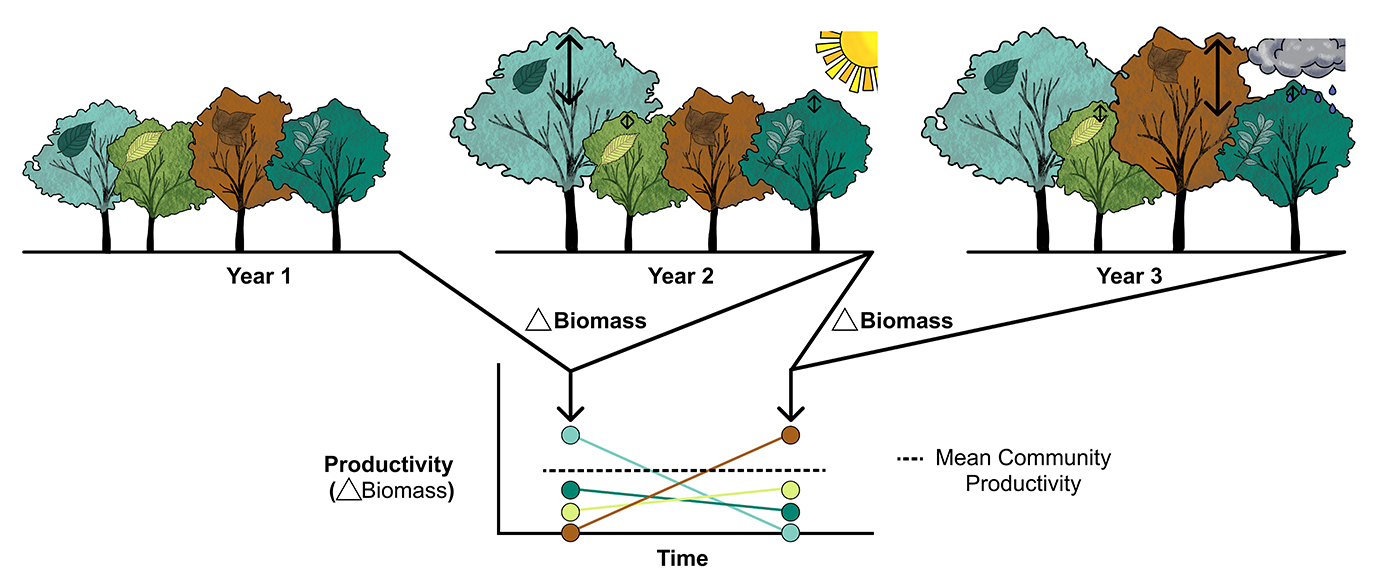
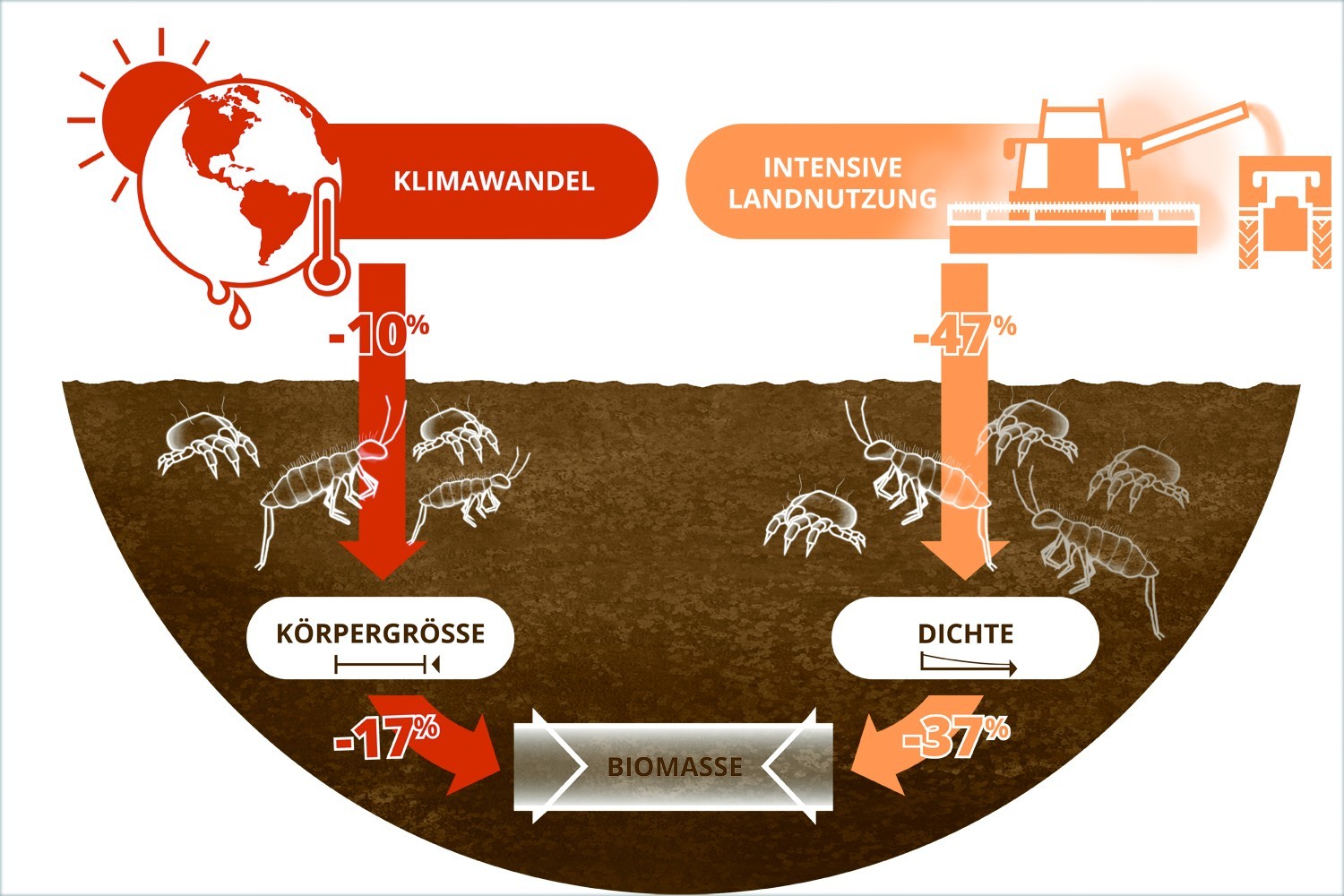
Nutrients strengthen link between precipitation and plant growth, study finds
Global analysis reveals a stronger biomass growth when key nutrients are not limiting.
More16.04.2025 | iDiv, iDiv Members, Research
Experiment in Leipzig’s floodplain forest: Using tree mortality to support oak regeneration
More11.04.2025 | iDiv, Media Release, Research, Theory in Biodiversity Science
“Internet of nature” helps researchers explore the web of life
More10.04.2025 | Blog Post
Striving for Gender Equity in Academia: Insights from the iDiv Female Scientists Initiative
More03.04.2025 | iDiv, Media Release
New podcast “Inside Biodiversity”
Inside Biodiversity is a new podcast hosted by iDiv.
More03.04.2025 | iDiv, Media Release, Research, Species Interaction Ecology
Dark Diversity Reveals Global Impoverishment of Natural Vegetation
Nature study shows that the potential occurrence of plant species is significantly higher
More27.03.2025 | iDiv, Media Release, Research, sDiv
Study shows global seed plant distribution over millions of years
More26.03.2025 | iDiv, Media Release, Research, Theory in Biodiversity Science
How elephants plan their journeys: New study reveals energy-saving strategies
More24.03.2025 | Experimental Interaction Ecology, iDiv Members, Media Release
Tree diversity helps reduce heat peaks in forests
More20.03.2025 | Experimental Interaction Ecology, Media Release, Research
Resource-efficient tree species grow faster under real conditions, study shows
More13.03.2025 | Biodiversity Synthesis, Media Release, Research
New study refutes habitat fragmentation claim
More11.03.2025 | Blog Post, iDiv, sDiv
Unraveling the Secrets of Grassland Biodiversity in Ukraine
How an sDiv collaboration helped reunite Ukrainian scholars scattered across Europe due to the war and provided them with an environment of stability, focus, and scientific exchange.
More05.03.2025 | iDiv, Media Release, Research
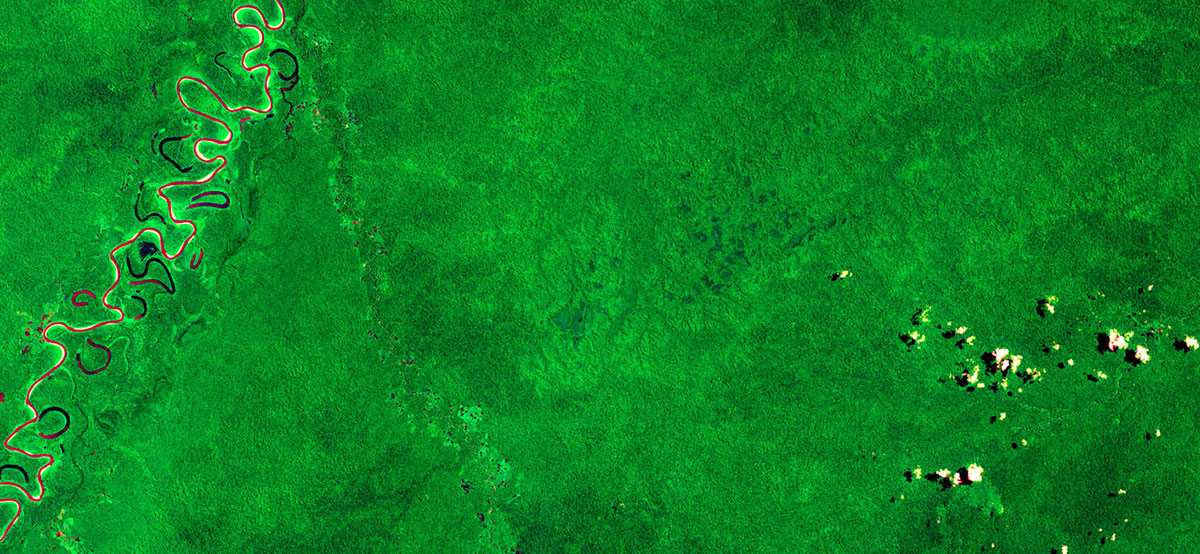
Satellite image analysis new insight into tropical forests
New Nature paper: satellite images from space are allowing scientists to delve deeper into the individual functions of different tropical forest canopies.
More05.03.2025 | Biodiversity Conservation, iDiv, Media Release
The future of nature conservation in Europe
An international research team is pioneering an approach to European conservation, integrating biodiversity as a solution to environmental challenges.
More27.02.2025 | Blog Post, sDiv
50,000 sDiv citations – 12 years of excellence in biodiversity research
More19.02.2025 | Experimental Interaction Ecology, iDiv, Media Release
Translating Soil Biodiversity receives Grüter Prize
The Werner and Inge Grüter Foundation’s 2025 Award for Science Communication goes to ‘Translating Soil Biodiversity’.
More13.02.2025 | Experimental Interaction Ecology, iDiv, Media Release, Research, sDiv
Hidden engineers help shape terrestrial ecosystems
A new paper in Nature shows how soil invertebrates’ engineering effects drive global ecosystem functions.
More
30.01.2025 | Experimental Interaction Ecology, iDiv, Media Release
Transfer prize for Translating Soil Biodiversity project
The team behind the ‘Translating Soil Biodiversity’ project at the German Centre for Integrative Biodiversity Research (iDiv) has been awarded the Transfer Prize of Leipzig University.
More
29.01.2025 | Biodiversity Synthesis, Media Release, Theory in Biodiversity Science
Climate change reshuffles species like a deck of cards
A new study found that biodiversity has changed faster in locations where warming or cooling was faster
More
15.01.2025 | iDiv Members, Media Release, Research
New technique by scientists reveals genetic ties within animal populations
More13.01.2025 | Biodiversity and People, iDiv, Media Release
Perspective: Tree crops key to advancing Sustainable Development Goals
More09.01.2025 | Biodiversity Conservation, iDiv Members, Media Release
Chimpanzees are genetically adapted local habitats and infections such as malaria
Chimpanzees bear genetic adaptations that help them thrive in their different forest and savannah habitats, some of which may protect against malaria
More
18.12.2024 | iDiv Members, Media Release, Soil Biodiversity and Functions
Anton Potapov receives ERC Consolidator Grant
The grant will fund the iDiv member’s research project CARBONWEB with around two million euros
More
04.12.2024 | Biodiversity Conservation, TOP NEWS
Henrique Pereira elected member of the Academia Europaea
Membership honours individuals who have demonstrated “sustained academic excellence”
More04.12.2024 | Experimental Interaction Ecology, Media Release, TOP NEWS
Survival Strategies of Tiny Soil Microbes Unravelled
New Nature paper has revealed how soil microbes are impacted by extreme weather events, offering new insights into the risks posed by climate change.
More03.12.2024 | iDiv Members, MLU News, TOP NEWS
Ecosystems: new study questions common assumption about biodiversity
Plant species can fulfil different functions within an ecosystem, even if they are closely related to each other.
More21.11.2024 | Science-Policy, TOP NEWS
iDiv researchers at the Convention on Biological Diversity’s COP16
Addressing biodiversity challenges with science-based strategies
More19.11.2024 | Experimental Interaction Ecology, Media Release, TOP NEWS, Unkategorisiert
Soil ecosystem more resilient when land managed extensively
Researchers studied the effects of intensive and extensive land use on soil biodiversity in cropland and grassland
More12.11.2024 | Media Release, TOP NEWS
Diverse and diverging demands on forests in Germany
Research team analyse biodiversity, ecosystems and economics of enriching beech forests with conifers
More
01.11.2024 | Biodiversity and People, TOP NEWS
Aletta Bonn appointed
iDiv researcher has been appointed to the German Advisory Council on Global Change
More21.10.2024 | Biodiversity and People, TOP NEWS
Scientific backing for the join-in campaign ”Unsere Flüsse“ on German television
Researchers investigate the habitat quality of Germany’s streams
More10.10.2024 | Biodiversity and People, TOP NEWS
Julia von Gönner wins Research Award for Citizen Science
The Citizen Science project FLOW mobilised over 900 citizens to contribute to the ecological monitoring of small streams.
More10.10.2024 | iDiv Members, Media Release, sDiv, TOP NEWS
European forest plants are migrating westwards, nitrogen main cause
Based on a press release by Ghent University
More08.10.2024 | sDiv, TOP NEWS
‘Life on the edge’: A new toolbox to predict global change impact on wildlife
New climate change prediction tool provides insight into population-level vulnerability to global change through combining genomic, geographic and environmental data.
More01.10.2024 | Media Release, Research, Science-Policy, TOP NEWS
Faktencheck Artenvielfalt shows the state of biodiversity in Germany
More than half of Germany’s habitat types have an ecologically unfavourable status
More01.10.2024 | iDiv, Media Release, TOP NEWS
New Phase for iDiv: Biodiversity Research Consolidated in Central Germany
Today, the German Centre for Integrative Biodiversity Research (iDiv) enters a new phase with new funding, a new COO, and a new research group.
More17.09.2024 | iDiv Members, Media Release, TOP NEWS
Pollen affects cloud formation and precipitation patterns
New study shows climate impact
More
16.09.2024 | Media Release, MLU News, TOP NEWS
Plants, Ecosystems and Climate Change: International Conference of the German Society for Plant Sciences in Halle
Botanik-Tagung takes place from 15 to 19 September at Martin Luther University Halle-Wittenberg
More10.09.2024 | iDiv Members, Media Release, TOP NEWS
Cloud cover reduced by large-scale deforestation
Serious impact on climate, say researchers
More09.09.2024 | Biodiversity Synthesis, Media Release, MLU News, TOP NEWS
Island life slows animals down
Birds and mammels on islands have a slower metabolism than their closest relatives on the mainland
More06.09.2024 | Biodiversity Economics, Media Release, TOP NEWS
Research Training Group ECO-N has started
Early career researchers study sustainable development
More27.08.2024 | Experimental Interaction Ecology, Media Release, Research, TOP NEWS
Environmental stressors weaken ecosystem resistance to change
The higher the environmental stress, the lower the resistance to global change.
More21.08.2024 | iDiv Members, Media Release, Research, TOP NEWS
Healthier honey bees found near organic fields and flower strips
A new study reveals how farming practices affect colony health.
More07.08.2024 | iDiv, Media Release, TOP NEWS
Unlocking the Wealth of Biodiversity Knowledge
Researchers developed a comprehensive framework to promote diversity, equity, and inclusion in global biodiversity research
More
22.07.2024 | Media Release, TOP NEWS, UFZ-News
Less productive yet more stable
Low-intensity grassland is better able to withstand the consequences of climate change
More19.07.2024 | Biodiversity and People, Research, TOP NEWS
Considering the mental health implications of nature conservation
Based on a joint text by Imperial College London and the University of Oxford
More09.07.2024 | iDiv Members, Media Release, Research, TOP NEWS
How a plant app helps identify the consequences of climate change
Leipzig. A research team led by the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University has developed an algorithm that analyses observational data from the Flora Incognita app. The novel approach described in Methods in Ecology and Evolution can be used to derive ecological patterns that could provide valuable information about the effects of climate change on plants.
More02.07.2024 | Biodiversity and People, Biodiversity Conservation, iDiv, Media Release, Science-Policy, TOP NEWS
What do we need for better biodiversity monitoring in Europe?
Based on a media release of IIASA A new publication in Conservation Letters authored by scientists from the German Centre for Integrative Biodiversity Research (iDiv) and the International Institute for Applied Systems Analysis (IIASA) with a large European consortium provides vital insights into the current status of biodiversity monitoring in Europe, identifying policy needs, challenges, and future pathways.
More28.06.2024 | Biodiversity Synthesis, Research, TOP NEWS
New study helps unravel the paradox of habitat fragmentation on biodiversity
Based on a media release by Professor Dr Jinbao Liao, Yunnan University Habitat fragmentation, a process where a large, continuous habitat is divided into smaller, isolated areas, can both help and harm biodiversity, depending on the total amount of habitat remaining in the landscape, according to a new study published in Nature Ecology & Evolution.
More26.06.2024 | Biodiversity Conservation, Media Release, TOP NEWS
Closing the Biodiversity Knowledge Gap: Unlocking Biodiversity Insights from the Tropical Andes
Report by Jose Valdez, postdoctoral researcher of Biodiversity Conservation at iDiv and Martin Luther University Halle-Wittenberg (MLU) Despite hosting some of the world’s most biodiverse ecosystems and the urgency of the region’s conservation challenges, researchers in Bolivia, Ecuador, and Peru often struggle to share their unique insights into these complex ecosystems with the global scientific community. This results in a “publication gap” where crucial biodiversity knowledge from the region remains underrepresented in global conversations.
More25.06.2024 | iDiv, Media Release, Science-Policy, TOP NEWS
Germany’s leading role in biodiversity research and policy
More17.06.2024 | Experimental Interaction Ecology, Media Release, TOP NEWS
Soil fauna has the potential to fundamentally alter carbon storage in soil
Based on a media release of the Czech Academy of Sciences The life strategies of a multitude of soil faunal taxa can strongly affect the formation of labile and stabilized organic matter in soil, with potential consequences for how soils are managed as carbon sinks, nutrient stores, or providers of food. This is the main conclusion of a review led by a team of researchers from the Czech Academy of Sciences, the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), and the Senckenberg Society for Nature Research. Based on a review of more than 180 scientific articles, the authors highlight major pathways by which soil fauna can influence soil organic matter stability, identify knowledge gaps, and suggest future research directions. The study has recently been published in Nature Communications.
More13.06.2024 | Biodiversity Economics, Experimental Interaction Ecology, Media Release, Physiological Diversity, TOP NEWS
Land management and climate change affect ecosystems’ ability to provide multiple services simultaneously
A novel study published in Nature Communications found that agroecosystems in Central Germany, specifically grasslands and croplands, may have an enhanced capacity to provide multiple goods and services simultaneously when land management reduces the use of pesticides and mineral nitrogen fertiliser.
More30.04.2024 | iDiv, Media Release, TOP NEWS
Low-intensity grazing is locally better for biodiversity but challenging for land users, a new study shows
A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), and the Helmholtz Centre for Environmental Research (UFZ) has investigated the motivation and potential incentives for and challenges of low-intensity grazing among farmers and land users in Europe. The interview results have been published in Land Use Policy.
More26.04.2024 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS
Climate change could become the main driver of biodiversity decline by mid-century
More23.04.2024 | iDiv Members, Media Release, Research, TOP NEWS
Study shows how plants regulate the climate in Europe
Based on a media release of Martin Luther University Halle-Wittenberg The climate regulates plant growth, but the climate is also regulated by plants. Depending on the vegetation composition, ecosystems even have a strong influence on the climate in Europe, a study by Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) in the journal Global Change Biology shows. The researchers linked satellite data with around 50,000 vegetation records from across Europe. A good five per cent of regional climate regulation can be explained by local plant diversity. The analysis also shows that the effects depend on many other factors. Plants regulate the climate by reflecting sunlight or cooling their surroundings through evaporation.
More18.04.2024 | Experimental Interaction Ecology, iDiv, Media Release, TOP NEWS
iDiv’s Nico Eisenhauer becomes member of the Leopoldina Academy
More17.04.2024 | Biodiversity Conservation, Media Release, TOP NEWS
Rewilding amphibians: Protecting endangered species to restore ecosystems
By Dr Gavin S. Stark, a postdoctoral researcher in the Biodiversity Conservation group at iDiv and lead author of the study.
More12.04.2024 | iDiv Members, TOP NEWS
iDiv’s founding director Christian Wirth Member of the Saxon Academy
More03.04.2024 | Biodiversity Conservation, iDiv, Media Release, Research, TOP NEWS
Demand for critical minerals puts African great apes at risk
A recent study led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the non-profit conservation organization Re:wild shows that the threat of mining to the great ape population in Africa has been greatly underestimated. Their results have been published in Science Advances.
More02.04.2024 | Media Release, TOP NEWS
iDiv is mourning the loss of Diana Wall
Diana Wall passed away on the 25th of March 2024. Diana Wall, a leading environmental scientist and soil ecologist, has been a longstanding member of iDiv’s Scientific Advisory Board (SAB) and part of several research endeavours at iDiv.
More28.03.2024 | Media Release, TOP NEWS
Genetic traces of hierarchy: How social standing shapes the epigenome of Tanzania’s Spotted Hyenas
Based on a media release from the Leibniz Institute for Zoo and Wildlife Research (Leibniz-IZW) A research consortium led by scientists from the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig and the Leibniz Institute for Zoo and Wildlife Research (Leibniz-IZW) provide evidence that social behaviour and social status are reflected at the molecular level of gene activation (epigenome) in juvenile and adult free-ranging spotted hyenas. The results, published in the scientific journal Communications Biology, contribute to a better understanding of the role of epigenetic mechanisms in the interplay of social, environmental and physiological factors in the life of a highly social mammal.
More26.03.2024 | Biodiversity Conservation, Media Release, MLU News, TOP NEWS
What do birds and rivers have to do with euro notes?
More14.03.2024 | Media Release, Research, Science-Policy, TOP NEWS
Citizen Science Project FLOW shows that Germany’s small streams are not in a good ecological state
More27.02.2024 | Media Release, Theory in Biodiversity Science, TOP NEWS
Extinctions could result as fish change foraging behaviour in response to rising temperatures
More23.02.2024 | Media Release, sDiv, TOP NEWS
Disentangling Nature’s Contributions to International Trade
Researchers have developed a multi-step process to quantify the dependency of international trade and so-called Nature’s contributions to people. With their new approach, which has been published in People and Nature, the researchers hope to improve knowledge about the complex relationship between nature and international trade.
More22.02.2024 | Biodiversity Synthesis, Research, TOP NEWS
Increasingly similar or different? Centuries-long analysis suggests biodiversity is differentiating and homogenising to a comparable extent
More20.02.2024 | iDiv, Media Release, TOP NEWS
Converting rainforest to plantation impacts food webs and biodiversity
Based on a media release of the University of Goettingen The conversion of rainforest into plantations erodes and restructures food webs and fundamentally changes the way these ecosystems function, according to a new study published in Nature. The findings provide the first insights into the processing of energy across soil and canopy animal communities in mega-biodiverse tropical ecosystems.
More14.02.2024 | iDiv, Media Release, Research, TOP NEWS
Critical Habitats at Risk: Three-quarters of vegetation types in the Americas are under-protected
The study published in Global Ecology and Conservation found that three-quarters of these distinct habitats in North, Central, and South America fall below the Global Biodiversity Convention’s target of 30% protection. The research led by the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) also highlights that over 40% of threatened bird and mammal species are mostly found in a single vegetation type, putting them at risk for extinction if these critical habitats remain unprotected.
More14.02.2024 | Biodiversity and People, Media Release, TOP NEWS
Fish in the upper Danube could be just as endangered in the future as they were in the past, but for different reasons
Based on a media release from the Leibniz Institute of Freshwater Ecology and Inland Fisheries (IGB) Rivers belong to the most threatened ecosystems on Earth. While many studies have projected climate change effects on species, little is known about the severity of these changes compared to historical alterations. Researchers led by the Leibniz Institute of Freshwater Ecology and Inland Fisheries (IGB) and the German Centre for Integrative Biodiversity Research (iDiv) explored the vulnerability of 48 native fish species in the upper Danube River Basin to past and potential future environmental changes. They show that fish have been particularly sensitive to changes in flow regimes in the past, while higher temperatures will pose the greatest threat in the future. The threat assessment will remain at least as high in the future. However, it could probably be mitigated by reconnecting former floodplains and improving river connectivity.
More09.02.2024 | Media Release, MLU News, TOP NEWS
Ecologist Brian McGill receives Humboldt Research Award
Based on a media release of Martin Luther University Halle-Wittenberg The Alexander von Humboldt Foundation has honoured US scientist Professor Dr Brian McGill from the University of Maine with the prestigious Humboldt Research Award. The biodiversity researcher was nominated by Professor Dr Jonathan Chase, head of Biodiversity Synthesis at the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU). The award is endowed with 60,000 euros. McGill will use the money for several research stays at iDiv.
More22.01.2024 | Biodiversity Conservation, iDiv, Media Release, Research, TOP NEWS
Wolves and elk are (mostly) welcome back in Poland and Germany’s Oder Delta region, survey shows
An online survey conducted in Germany and Poland shows that large parts of the participants support the return of large carnivores and herbivores, such as wolves and elk, to the Oder Delta region, according to a study published in People and Nature. Presented with different rewilding scenarios, the majority of survey participants showed a preference for land management that leads to the comeback of nature to the most natural state possible. Locals, on the other hand, showed some reservations.
More22.01.2024 | Media Release
Bloom or bust
A new white paper ‘Bloom or bust’, that has been produced by USB together with experts including Dr Miguel Fernandez from the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg, takes stock of where the world stands on biodiversity loss, existing solutions, and the role that finance, government action, and collaboration can play.
More17.01.2024 | iDiv Members, Media Release, TOP NEWS
Honoured: Christian Wirth appointed External Member of the Max Planck Institute for Biogeochemistry
Based on a media relase by the Max Planck Institute for BiogeochemistryThe Max Planck Institute for Biogeochemistry (MPI BGC) has gained a new external member: Prof Dr Christian Wirth has been appointed by the Senate of the Max Planck Society as External Scientific Member at the request of the MPI BGC. Wirth is the founding director and spokesperson of the German Centre for Integrative Biodiversity Research (iDiv), professor at Leipzig University and director of the Leipzig Botanical Garden. The plant ecologist investigates the effects of natural and man-made changes in plant biodiversity on ecosystem processes, such as carbon storage, water consumption and energy balance. As a former group leader and later fellow at the MPI BGC, Wirth initiated and supported the development of the TRY database, the world’s largest collection on plant traits. His research on the influence of climate change on biodiversity and ecosystem services of forests ideally complements the research focus of the Max Planck Institute in Jena.
More09.01.2024 | Media Release, TOP NEWS
Global study of extreme drought impacts on grasslands and shrublands
Based on a media release of Colorado State University A global study organized and led by Colorado State University scientists and with participation of several researchers from the German Center for Integraitive Biodiversity Research (iDiv) shows that the effects of extreme drought – which is expected to increase in frequency with climate change – have been greatly underestimated for grasslands and shrublands.
More08.01.2024 | iDiv Members, Media Release, Research, TOP NEWS
Use of habitat for agricultural purposes puts primate infants at risk
Frequent visits to oil palm plantations are leading to a sharp increase in mortality rates among infant southern pig-tailed macaques (Macaca nemestrina) in the wild, according to a new study published in Current Biology. In addition to increased risk from predators and human encounters, exposure to harmful agricultural chemicals in this environment may negatively affect infant development.
More05.01.2024 | Biodiversity Economics, Media Release, Science-Policy, TOP NEWS
Environmental economist: “There is no justification for subsidising agricultural diesel”
More15.12.2023 | Media Release, TOP NEWS
How can Europe restore its nature?
Based on a media release of the University of Duisburg-Essen Early 2024, the European Parliament will take a final vote on the ‘Nature Restoration Law’ (NRL), a globally unique but hotly debated regulation that aims to halt and reverse biodiversity loss in Europe. An international team of scientists led by the University of Duisburg-Essen und with contribution from reseachers from the German Center for Integrative Biodiversity Research (iDiv) as well as the Helmholtz Centre for Environmental Research (UFZ) investigated the prospects of the new regulation. The article has been published in the Science Magazine.
More15.12.2023 | Biodiversity and People, Biodiversity Synthesis, Media Release, TOP NEWS
Common insect species are suffering the biggest losses
Leipzig. Insect decline is being driven by losses among the locally more common species, according to a new study published in Nature. Led by researchers at the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU), the meta-analysis of 923 locations around the world notes two significant trends: 1) the species with the most individuals (the highest abundance) are disproportionately decreasing in number, and 2) no other species have increased to the high numbers previously seen. This likely explains the frequent observation that there are fewer insects around now than ten, twenty, or thirty years ago.
More13.12.2023 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
How forests smell – a risk for the climate?
Based on a media release of Leipzig University Plants emit odours for a variety of reasons, such as to communicate with each other, to deter herbivores or to respond to changing environmental conditions. An interdisciplinary team of researchers from Leipzig University, the Leibniz Institute for Tropospheric Research (TROPOS) and the German Centre for Integrative Biodiversity Research (iDiv) carried out a study to investigate how biodiversity influences the emission of these substances. For the first time, they were able to show that species-rich forests emit less of these gases into the atmosphere than monocultures. In the past, it was assumed that forests with more species would release more emissions. Experiments by the Leipzig team have now disproved this assumption. Their study has been published in the journal Communications Earth & Environment.
More07.12.2023 | iDiv, Media Release, TOP NEWS
Recommendations for a research agenda on biodiversity and pandemics
Leipzig. An Expert Working Group with involvement of the German Centre for Integrative Biodiversity Research (iDiv) has published science-policy recommendations for future research to support transformative pandemic-prevention and preparedness policies. The report was published by Eklipse, a spinoff of a UFZ-led EU project.
More06.12.2023 | Experimental Interaction Ecology, iDiv, Media Release, Research, TOP NEWS
Deciphering nature’s climate shield: Plant diversity stabilises soil temperature
Based on a media release of Leipzig University A new study has revealed a natural solution to mitigate the effects of climate change, such as extreme weather events. Researchers from Leipzig University, the Friedrich Schiller University Jena, the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig (iDiv) and other research institutions have discovered that high plant diversity acts as a buffer against fluctuations in soil temperature. This buffer can then be of vital importance to ecosystem processes. They have just published their new findings in the journal Nature Geoscience.
More04.12.2023 | iDiv, Media Release, TOP NEWS
Ecology: “iDiv Universities” climb to the top of the international Shanghai Ranking 2023
Halle, Jena, Leipzig. The three universities that comprise the German Centre for Integrative Biodiversity Research’s (iDiv) consortium rose to the top of this year’s international Shanghai Ranking in the field of ecology. Of the 5,000 universities evaluated, the universities of Halle, Jena, and Leipzig placed 27th, 51st*, and 35th, respectively. This recognition means Central Germany is home to three of the country’s six highest-ranked universities in the field.
More15.11.2023 | TOP NEWS
Highly Cited Researchers 2023
Clarivate Analytics lists eight iDiv members in its 2023 selection of “Highly Cited Researchers”. According to Clarivate Analytics, these scientists have demonstrated significant influence through the publication of multiple papers, highly cited by their peers, during the last decade.
More13.11.2023 | Biodiversity Economics, iDiv, Media Release, TOP NEWS
DFG gives green light for new Research Training Group
Based on a media release of Leipzig University Another great success for Leipzig in supporting early career researchers: the German Research Foundation (DFG) announced that it will provide around 7 million euros in funding for a new Research Training Group from April 2024. Under the leadership of Professor Martin Quaas, head of Biodiversity Economics at the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University, more than 40 doctoral researchers in the fields of economics and management science, the natural and life sciences will investigate sustainable concepts for the use of natural common goods.
More13.11.2023 | iDiv, Media Release, TOP NEWS
When writers write about biodiversity
Based on a media release of Leipzig University Novels and poems often contain descriptions of plants or animals – sometimes more, sometimes less detailed. The extent to which flora and fauna feature in a literary work also depends on who wrote it and under what circumstances. For example, female authors tend to use more species names when they write. This is the conclusion of a research team from Leipzig University, the German Centre for Integrative Biodiversity Research (iDiv) and Goethe University Frankfurt, who examined around 13,500 literary works by approximately 2,900 authors. The study published in People and Nature is an example of how methods from the natural sciences and the humanities can be combined using digital techniques.
More03.11.2023 | Biodiversity Conservation, TOP NEWS
Tailoring Biodiversity Information to Local Needs in the Threatened Tropical Andes
Lima, Halle, Leipzig. Sustainable biodiversity conservation requires cooperation among scientific, societal, economic, and political institutions. In the journal Conservation Science and Practice, researchers have published a new approach to co-designing biodiversity indicators relevant for conservation. They brought together multiple stakeholders in a consultative process, tailoring user-relevant biodiversity information to local needs. The project was led by researchers of the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg, together with multiple partners in the Tropical Andes. The collaborative approach can serve as a blueprint for making biodiversity information more inclusive, considering the diverse worldviews, values, and knowledge systems between science, policy, and practice.
More30.10.2023 | Experimental Interaction Ecology, Media Release, Theory in Biodiversity Science, TOP NEWS
Even low levels of artificial light disrupt ecosystems
Leipzig, Jena. A new collection of papers on artificial light at night show the impact of light pollution to be surprisingly far-reaching, with even low levels of artificial light disrupting species communities and entire ecosystems. Published in Philosophical Transactions of the Royal Society B, the special theme issue, which includes 16 scientific papers, looks at the effects of light pollution in complex ecological systems, including soil, grassland, and insect communities. Led by researchers at the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich Schiller University Jena, the collection notes the increasing ubiquity of light pollution, while emphasising the domino effect light pollution has on ecosystem function and stability.
More27.10.2023 | Biodiversity and People, iDiv, Media Release, Research, TOP NEWS
How social media can contribute to species conservation
Leipzig. Photos of plant and animal species that are posted on social media can help protect biodiversity, especially in tropical regions. This is the conclusion of a team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ), the Friedrich Schiller University Jena (FSU), and the University of Queensland (UQ). Recently published in BioScience, One Earth, and Conservation Biology, the three studies investigated the benefits of using Facebook data for conservation assessments in Bangladesh. The researchers point out that social media can support species monitoring and significantly contribute to conservation assessments in tropical countries.
More10.10.2023 | Experimental Interaction Ecology, Media Release, TOP NEWS
Funding new research in the Jena Experiment – focus on ecosystem stability
Joint media release from iDiv, Leipzig University and Friedrich Schiller University Jena Jena/Leipzig. The Deutsche Forschungsgemeinschaft (DFG) is to fund a Research Unit in the Jena Experiment for a further four years with around five million euros. The scientists, led by the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University and the Friedrich Schiller University Jena, will focus in particular on the stabilising effect of biodiversity against extreme climate events such as drought, heat and frost.
More09.10.2023 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
Mixed forests are more productive when they are structurally complex
Dresden, Halle, Leipzig. Tree species richness increases forest productivity by enhancing aboveground structural complexity. This is the key result of a joint study by TUD Dresden University of Technology, the German Centre for Integrative Biodiversity Research (iDiv), Leuphana University Lüneburg, Martin Luther University Halle-Wittenberg, Leipzig University, and the University of Montpellier. The results have been published in the journal Science Advances.
More26.09.2023 | Experimental Interaction Ecology, iDiv, Media Release, Research, TOP NEWS
Invertebrate decline reduces natural pest control and decomposition of organic matter
Leipzig. The decline in invertebrates also affects the functioning of ecosystems, including two critical ecosystem services: aboveground pest control and belowground decomposition of organic material, according to a new study published in Current Biology and led by researchers at the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University. The study provides evidence that loss of invertebrates leads to a reduction in important ecosystem services and to the decoupling of ecosystem processes, making immediate protection measures necessary.
More08.09.2023 | Biodiversity Synthesis, iDiv, Research, TOP NEWS
Big fish are shrinking and small fish are multiplying, a new study shows
Based on a media release from the University of St. Andrews St. Andrews/Leipzig. Organisms are becoming smaller through a combination of species replacement, and changes within species. Published in Science, the research looked at time series covering the past 60 years, from many types of animals and plants around the world. The study was conducted by an international team of scientists from 17 universities, as part of a working group funded by the German Centre for Integrative Biodiversity Research (iDiv), and led by scientists of the University of St Andrews, and the University of Nottingham.
More08.09.2023 | iDiv, Media Release, TOP NEWS
iDiv’s early career researchers’ response to the proposed changes to the WissZeitVG
iDiv’s researchers1 are concerned about the already precarious working conditions in German academia and are alarmed that the proposed changes to the WissZeitVG will exacerbate this.
More04.09.2023 | Biodiversity Conservation, Biodiversity Synthesis, Media Release, TOP NEWS
Most species are rare. But not very rare
Halle/Saale, Fort Lauderdale. More than 100 years of observations in nature have revealed a universal pattern of species abundances: Most species are rare but not very rare, and only a few species are very common. These so-called global species abundance distributions have become fully unveiled for some well-monitored species groups, such as birds. For other species groups, such as insects, however, the veil remains partially unlifted. These are the findings of an international team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the University of Florida (UF), published in the journal Nature Ecology and Evolution. The study demonstrates how important biodiversity monitoring is for detecting species abundances on planet Earth and for understanding how they change.
More01.09.2023 | iDiv, TOP NEWS
New map on potentially groundwater-dependent vegetation in the Mediterranean biome
Report by Léonard El-Hokayem, doctoral student at Martin Luther University Halle-Wittenberg and the German Centre for Integrative Biodiversity Research (iDiv) Halle. Decreasing rainfall and increased groundwater use are threatening vegetation and ultimately biodiversity in the Mediterranean biome. Plants that depend on groundwater are particularly vulnerable. We have developed a novel, easy-to-use index to map potentially groundwater dependent vegetation (pGDV) based on environmental site conditions and vegetation characteristics. Our concept combines globally-available geodata and remote sensing and has recently been published in Science of The Total Environment. The results indicate that 31 % of the natural vegetation in the Mediterranean likely depends on groundwater. A biome-wise map of pGDV is important to prioritise areas for detailed identification of actual GDV and biodiversity conservation.
More25.08.2023 | iDiv, Media Release, TOP NEWS
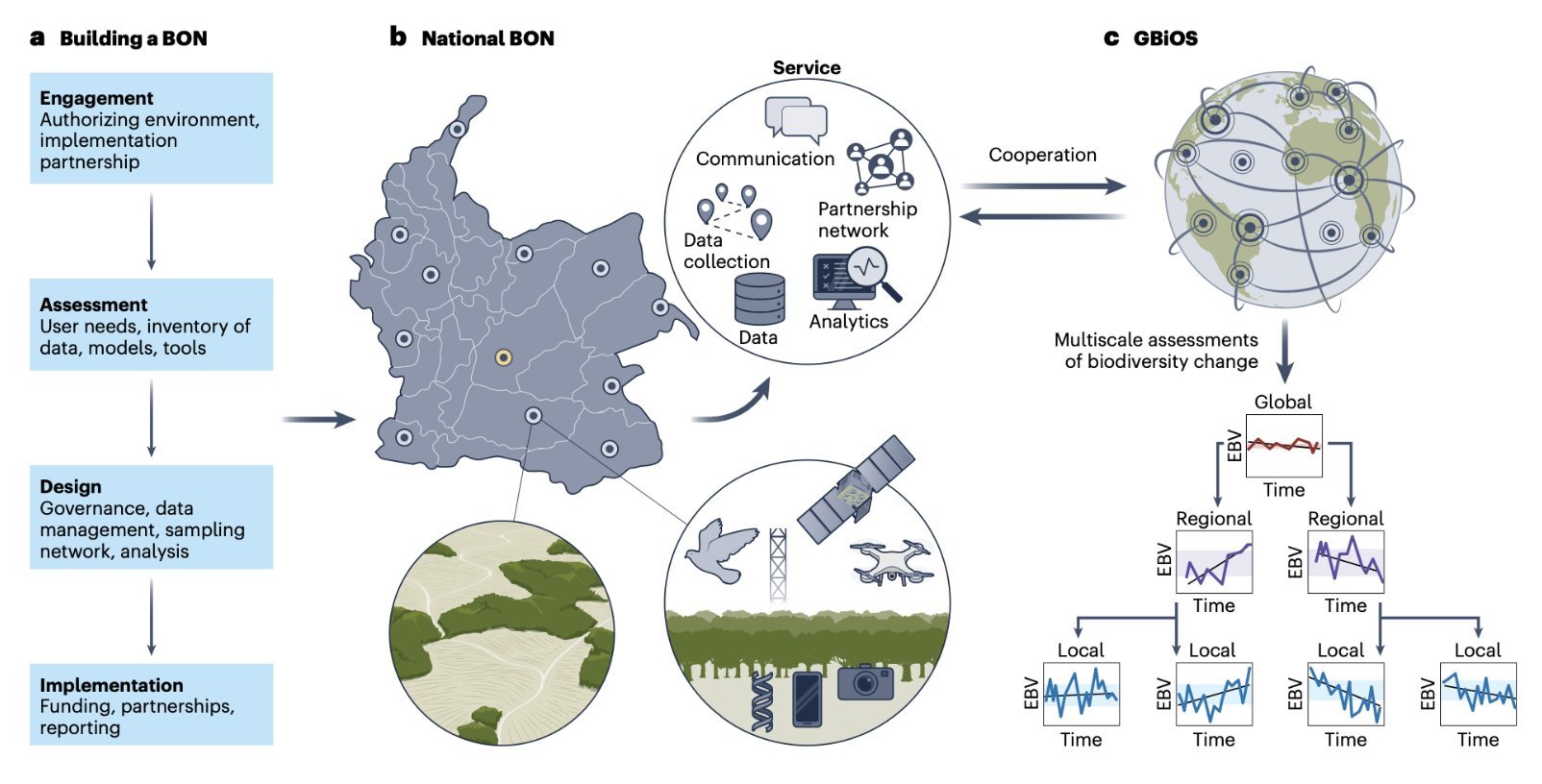
A global observatory to monitor Earth’s biodiversity
Based on a media release of GEO BON The Global Biodiversity Observing System (GBiOS) is a proposal developed by scientists from the Group on Earth Observations Biodiversity Observation Network (GEO BON), and its partners, including the German Center for Integrative Biodiversity Research (iDiv). It will combine technology, data, and knowledge from around the world to foster collaboration and data sharing among countries and to provide the data urgently needed to monitor biodiversity change and target action. The proposal for this novel system was published in Nature Ecology & Evolution.
More23.08.2023 | iDiv, iDiv Members, Media Release, Research, TOP NEWS
1000 new species for Nigeria
Leipzig/Katsina. To date, over 1000 vascular plants in Nigeria may be undescribed, making it impossible to know whether or not these plants are endangered and in need of protection. This is one of the key results of a new study led by researchers from the German Center for Integrative Biodiversity Research (iDiv) and Leipzig University, published in Annals of Botany. In order to meet the targets of the CBD’s Global Biodiversity Framework, urgent measures are required that promote local taxonomic activities.
More15.08.2023 | Media Release, TOP NEWS
Some plants do not shed their leaves in autumn, for good reason
Report by Dr Gerrit Angst, postdoctoral researcher of the Experimental Interaction Ecology at iDiv, Leipzig University, and the Czech Academy of Sciences and co-author: Leipzig/Budweis/Prague. Retention of dead biomass by plants is common in the temperate herbaceous flora and can be related to certain plant traits, indicating relevance to ecosystem functioning. These are the main findings of an experimental study on more than 100 plant species jointly performed by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University, the Czech Academy of Sciences, and the Charles University, Prague. The study has recently been published in the Journal of Ecology.
More08.08.2023 | Media Release, sDiv, TOP NEWS
Satellite documentation of effects of heat waves on plants
More12.07.2023 | iDiv, Media Release, TOP NEWS
Remote plant worlds
Based on a media release by the University of Göttingen Oceanic islands provide useful models for ecology, biogeography and evolutionary research. Many ground-breaking findings – including Darwin’s theory of evolution – have emerged from the study of species on islands and their interplay with their living and non-living environment. Now, an international research team led by researchers from the University of Göttingen and the German Centre for Integrative Biodiversity Research (iDiv) has investigated the flora of the Canary Island of Tenerife. The results were surprising: the island’s plant-life exhibits a remarkable diversity of forms. However, the plants differ little from mainland plants in functional terms. However, unlike the flora of the mainland, the flora of Tenerife is dominated by slow-growing, woody shrubs with a “low-risk” life strategy. The results were published in Nature.
More15.06.2023 | Experimental Interaction Ecology, Media Release, TOP NEWS
Holistic management is key to increase carbon sequestration in soils
Report by Dr Gerrit Angst, postdoctoral researcher of the Experimental Interaction Ecology at iDiv, Leipzig University, and the Czech Academy of Sciences and first author: Leipzig/Budweis/Copenhagen. Increased carbon sequestration in soil to help mitigate climate change can only be achieved by a more holistic management. This is the conclusion from our opinion paper conceptualized together with colleagues from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University, the Czech Academy of Sciences, and the University of Copenhagen. We developed a novel framework that can guide informed and effective management of soils as carbon sinks. The study has recently been published in Nature Communications.
More26.04.2023 | iDiv, Media Release, Research, Theory in Biodiversity Science, TOP NEWS
Diverse landscapes help insects cope with heat stress
Leipzig/Jena/Bad Lauchstädt. Global warming is affecting terrestrial insects in multiple ways. In response to increasingly frequent heat extremes, they have to either reduce their activity or seek shelter in more suitable microhabitats. A new study led by researchers from the German Centre for Integrative Biodiversity Research (iDiv) and Friedrich Schiller University Jena shows: The more diverse these microhabitats are, the better for the insects. For their study, published in Global Change Biology, they developed a new approach to accurately track insect movements and activity.
More20.04.2023 | iDiv, Media Release, TOP NEWS
iDiv celebrates its 10th anniversary
The German Centre for Integrative Biodiversity Research (iDiv) celebrated its 10th anniversary today with a ceremony in the Leipzig University Paulinum. Over 300 guests from politics, science and civil society took part, including the ministers-president of Saxony, Saxony-Anhalt and Thuringia, Michael Kretschmer, Dr Reiner Haseloff and Bodo Ramelow as well as the Federal Government Commissioner for eastern Germany, Carsten Schneider. They acknowledged the research centre’s important contributions to the protection of biological diversity. In his welcoming message, Federal Chancellor Olaf Scholz stressed the importance of “world-class basic research” for international biodiversity policy.
More19.04.2023 | iDiv, Media Release, Research, Theory in Biodiversity Science, TOP NEWS
Large animals travel more slowly because they can’t keep cool
Leipzig. Whether an animal is flying, running or swimming, its traveling speed is limited by how effectively it sheds the excess heat generated by its muscles, according to a new study led by Alexander Dyer from the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich Schiller University Jena, published in the open access journal PLOS Biology.
More30.03.2023 | Biodiversity Synthesis, iDiv, Media Release, TOP NEWS
New ideas for biodiversity research: ecologist Jonathan Chase receives ERC Advanced Grant
Joint media release of the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg The European Research Council (ERC) announced that Prof. Dr. Jonathan Chase will be awarded one of the prestigious ERC Advanced Grants. The scientist will receive almost 2.5 million euros over the next five years to fund his research project “MetaChange”. With this project, he plans to develop new concepts, tools and analyses for a better understanding of biodiversity and its change. Chase has been conducting research and teaching at the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) since 2014.
More20.03.2023 | Biodiversity Synthesis, iDiv, Media Release, Research, TOP NEWS
Widespread species gaining ground
Leipzig/Halle. Human activities are accelerating biodiversity change and promoting a rapid turnover in species composition. A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) has now shown that more widespread species tend to benefit from anthropogenic changes and increase the number of sites they occupy, whereas more narrowly distributed species decrease. Their results, which were published in Nature Communications, are based on an extensive dataset of over 200 studies and provide evidence that habitat protection can mitigate some effects of biodiversity change and reduce the systematic decrease of small-ranged species.
More16.03.2023 | Biodiversity Economics, iDiv, Media Release, Research, TOP NEWS
How fishermen benefit from reversing evolution of cod
Leipzig. Intense fishing and overexploitation have led to evolutionary changes in fish stocks like cod, reducing both their productivity and value on the market. These changes can be reversed by more sustainable and far-sighted fisheries management. The new study by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University and the Institute of Marine Research in Tromsø, which was published in Nature Sustainability, shows that reversal of evolutionary change would only slightly reduce the profit of fishing, but would help regain and conserve natural genetic diversity.
More09.03.2023 | Biodiversity Synthesis, iDiv, Media Release, TOP NEWS
Dwarfs and giants on islands more likely to go extinct
Leipzig/Halle. Islands are “laboratories of evolution” and home to animal species with many unique features, including dwarfs that evolved to very small sizes compared to their mainland relatives, and giants that evolved to large sizes. A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) has now found that species that evolved to more extreme body sizes compared to their mainland relatives have a higher risk of extinction than those that evolved to less extreme sizes. Their study, which was published in Science, also shows that extinction rates of mammals on islands worldwide increased significantly after the arrival of modern humans.
More09.03.2023 | iDiv Members, Media Release, MLU News, Research, TOP NEWS
Global climate data insufficiently explain composition of local plant species
Based on a media release of Martin Luther University Halle-Wittenberg The global climate influences regional plant growth – but not to the same extent in all habitats. This finding was made by geobotanists at Martin Luther University Halle-Wittenberg (MLU) and members of the German Centre for Integrative Biodiversity Research (iDiv) after analysing over 300,000 European vegetation plots. Their conclusion: No general prediction can be made about the effects of climate change on the Earth’s vegetation; instead, the effects depend to a large degree on local conditions and the habitat under investigation. The findings were published in the renowned journal Nature Communications.
More07.03.2023 | Media Release, TOP NEWS
Plant roots fuel tropical soil animal communities
Based on a media release by the University of Göttingen Göttingen/Leipzig. Soil animal communities in the tropics are driven by plant roots and the resources derived from them. This is the main finding of a new study of a research team led by the University of Göttingen, the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University. Millions of small creatures toiling in a single hectare of soil including earthworms, springtails, mites, insects, and other arthropods are crucial for decomposition and soil health. For a long time, it has been believed that leaf litter is the primary resource for these animals. However, this recent study published in the journal Ecology Letters shows that litter doesn’t play any crucial role at all for the tropical soil fauna.
More01.03.2023 | Biodiversity Conservation, Media Release, TOP NEWS
European conservation leaders gather to boost collective dialogue for a Trans-European Nature Network
Based on a media release by the International Institute for Applied Systems Analysis (IIASA) Brussels, 28 February 2023. More than 70 leading EU policy and governmental decision-makers joined to lay the foundation for a bold new vision for Europe’s nature protection in the first NaturaConnect Stakeholder Event this year. Organised by the Horizon Europe NaturaConnect project, the event welcomed a diverse range of influential stakeholders, from country representatives to European Union delegates and international and European conservation organisations. At the heart of the NaturaConnect project is the goal of supporting the creation of a Trans-European Nature Network (TEN-N) of protected and connected areas that conserve at least 30% of land in the EU and benefit both nature and people. The new project is conducted by international partners from research and environmental organisations, led by the International Institute for Applied Systems Analysis (IIASA), the German Centre for Integrative Biodiversity Research (iDiv) and the Martin-Luther University Halle-Wittenberg (MLU).
More10.02.2023 | Biodiversity Conservation, Media Release, TOP NEWS
How does biodiversity change globally? Detecting accurate trends may be currently unfeasible
Leipzig/Halle. Existing data are too biased to provide a reliable picture of the global average of local species richness trends. This is the conclusion of an international research team led by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU). The authors recommend prioritising local and regional assessments of biodiversity change instead of attempting to quantify global change and advocate standardised monitoring programmes, supported by models that take measurement errors and spatial biases into account. The study was published in the journal Ecography.
More09.02.2023 | Media Release, Molecular Interaction Ecology, Physiological Diversity, TOP NEWS
How could we evolve such a huge brain?
Based on a media release by the University of Amsterdam Amsterdam/Leipzig/Jena. A new study, published in the journal Frontiers in Ecology and Evolution, investigated the foraging behaviour of children in a present-day forager society. Already from an early age, there was a gender-specific development of foraging skills. These new findings, combined with the high level of food sharing in forager societies, support the embodied capital theory, offering an explanation for the substantially larger brains in humans. Foraging skills could have provided humans with a more stable energy and nutrient supply, which may ultimately have enabled large resource investments into the brain. The research was led by the University of Amsterdam (UvA), the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology, the Helmholtz Centre for Environmental Research (UFZ), and the Friedrich Schiller University Jena.
More07.02.2023 | Biodiversity Synthesis, Media Release, Physiological Diversity, TOP NEWS
Plant diversity may never fully recover from agriculture without a helping hand
Leipzig/Minnesota. Agriculture is considered a major disturbance for ecological systems – the recovery of degraded or formally used agricultural land might take a long time. However, without any active restoration interventions, this recovery can take an exceedingly long time and is often incomplete, as shown by a team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ). Their study, which was published in the Journal of Ecology, sheds light on the recovery process at different scales in former agricultural sites, pointing to specific restoration interventions that could help biodiversity to recovery.
More01.02.2023 | Biodiversity and People, Media Release, TOP NEWS
76 per cent of assessed insect species not adequately covered by protected areas
Leipzig/Jena. Insect numbers have been declining over the past decades in many parts of the world. Protected areas could safeguard threatened insects, but a team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ), the Friedrich Schiller University Jena and the University of Queensland now found that 76 per cent of globally assessed insect species are not adequately covered by protected areas worldwide. In the journal One Earth, the researchers encourage decision-makers to give more consideration to the by far biggest species group in implementing the new goals of the UN Convention on Biological Diversity.
More17.01.2023 | iDiv, Media Release, TOP NEWS
Implementing global biodiversity targets in Germany – with support from research
Berlin. Implementation of the recently agreed UN nature conservation targets will only succeed if all stakeholders – from policy and practice, civil society, business and science – work together. Science provides the knowledge base for effective action, thus playing a key role in the implementation of these global objectives. This was the tenor of discussion at the parliamentary evening on 17 January in the State Representation of Saxony-Anhalt in Berlin, to which the German Centre for Integrative Biodiversity Research (iDiv) and the Saxony-Anhalt Ministry of Science, Energy, Climate Protection and the Environment had invited. The scientists call for a high-level national biodiversity council as an essential element in making the conservation of biological diversity a core political issue across all relevant ministries.
More13.01.2023 | Experimental Interaction Ecology, iDiv, Media Release, Research, TOP NEWS
Grassland ecosystems become more resilient with age
Based on a media release of the University of Zurich Zurich/Leipzig. Recent experiments have shown that the loss of species from a plant community can reduce ecosystem functions and services such as productivity, carbon storage and soil health. With the loss of functioning the ecosystem may also become destabilized in its ability to maintain ecosystem functions and services in the long-term. However, assessing this is only possible if experiments can be maintained for a sufficient length of time.
More03.01.2023 | Media Release, Species Interaction Ecology, TOP NEWS, UFZ-News
Fewer moths, more flies
In the far north of the planet, climate change is clearly noticeable. A new study in Finland now shows that in parallel there have been dramatic changes in pollinating insects. Researchers from the Martin Luther University Halle-Wittenberg (MLU), the Helmholtz Centre for Environmental Research (UFZ), and the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig (iDiv) have discovered that the network of plants and their pollinators there has changed considerably since the end of the 19th century. As the scientists warn in an article published in Nature Ecology & Evolution, this could lead to plants being pollinated less effectively. This, in turn, would adversely affect their reproduction.
More02.01.2023 | Biodiversity Conservation, TOP NEWS
wildE: Restoring wild habitats in Europe against climate change
Based on a media release by INRAE Cestas/Halle. Terrestrial ecosystems throughout Europe face the twin threats of climate change and the loss of biodiversity. “Rewilding” could be an important ecological restoration solution to mitigate both of these issues, but up to now, it is mostly restricted to local initiatives scattered across the continent focussing on biodiversity objectives alone. The wildE project aims to assess the synergies between climate change mitigation, adaptation and biodiversity and thus to improve the potential of climate-smart rewilding as a nature-based solution for ecological restoration in Europe. wildE is funded by the EU Horizon programme and coordinated by the French National Research Institute for Agriculture, Food and Environment INRAE with the participation of the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg.
More15.12.2022 | iDiv Members, TOP NEWS
Identification of groundwater-dependent vegetation via satellites
Report by Léonard El-Hokayem (doctoral researcher at MLU and iDiv) Halle. Groundwater in (semi)-arid regions plays a key role in sustaining important terrestrial ecosystems, providing drinking water and supporting agriculture. We developed a new multi-instrument framework to identify groundwater-dependent vegetation (GDV) in these regions. Therefore, a combination of satellite images and other environmental data got tested and validated in a Mediterranean study area in Southern Italy. The developed concept was recently published in Ecological Indicators. It allows for the identification and study of vegetation that relies on groundwater, and thus can help protect these biodiversity hotspots and the ecosystem services they provide.
More14.12.2022 | Macroecology and Society, Media Release, sDiv, TOP NEWS
Humans and nature: The distance is growing
Joint media release by iDiv and Leipzig University Leipzig/Moulis. Humans are living further and further away from nature, leading to a decline in the number of our interactions with nature. This is the finding of a meta-study conducted by a Franco-German research team at the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University and the Theoretical and Experimental Ecology Station (SETE – CNRS). The researchers highlight that human experience of nature is crucial for developing pro-environmental behaviour and thus facing the global environmental crisis. The study has been published in Frontiers in Ecology and the Environment.
More12.12.2022 | Biodiversity Conservation, iDiv, Media Release, TOP NEWS
A novel and efficient method for monitoring Asian otters
South-east Asia is a melting pot of otters that are very difficult to monitor in the wild. We developed and tested a novel, accurate, and affordable DNA-based method to reliably identify three endemic Asian otter species in a paper published today in Ecology and Evolution. The protocol developed by our team of South Asian researchers from the Malaysian Nature Society, Sunway University, Department of Wildlife and National Parks (PERHILITAN), Peninsular Malaysia, Martin Luther University Halle-Wittenberg, and the German Centre for Integrative Biodiversity Research (iDiv) will help in monitoring and conservation of threatened otter species in Asia. We also highlight the importance and need of cost-effective and replicable techniques to advance biodiversity monitoring in highly biodiverse yet under-represented parts of the world.
More08.12.2022 | Biodiversity and People, iDiv, Media Release, Research, TOP NEWS, UFZ-News
Valid Data from Citizen Science
More01.12.2022 | TOP NEWS
Award for Prof. Walter Rosenthal
Jena. The President of the Friedrich Schiller University Jena, Prof. Walter Rosenthal, has been named “University Manager of the Year 2022”. Rosenthal has been Chair of the iDiv Board of Trustees since 2018.
More30.11.2022 | iDiv, iDiv Members, TOP NEWS
Biodiversity under climate extremes: Patient and Healer
Leipzig. The world is experiencing two megatrends: Climate extremes are increasing in magnitude and frequency while biodiversity is declining. Writing in Nature, researchers from Leipzig University and the German Centre for Integrative Biodiversity Research (iDiv) with collaborators across Europe, express concern that these two trends can mutually exacerbate each other. The researchers call for a new research agenda emphasizing the interconnecting risks of climate extremes and biodiversity decline.
More27.11.2022 | iDiv Members, Media Release, TOP NEWS
New Senckenberg Institute to be established in Jena
More24.11.2022 | iDiv, Research, TOP NEWS
Increased grazing pressure threatens the most arid rangelands
Based on a media release of the University of Alicante Grazing can have positive effects on ecosystem services, particularly in species-rich rangelands. However, these effects turn to negative under a warmer climate. This was found by a team of researchers led by the University of Alicante (UA) and with participation of the German Centre for Integrative Biodiversity Research (iDiv). The study, which was published in Science, reports results from the first-ever global field assessment of the ecological impacts of grazing in drylands.
More15.11.2022 | TOP NEWS
Highly Cited Researchers 2022
Clarivate Analytics lists seven iDiv members in its 2022 selection of “Highly Cited Researchers”. According to Clarivate Analytics, these scientists have demonstrated significant influence through the publication of multiple papers, highly cited by their peers, during the last decade.
More10.11.2022 | iDiv Members, Media Release, TOP NEWS
Deforestation and grassland conversion are the biggest causes of biodiversity loss
Based on a media release by Natural History Museum London Luxembourg/London/Halle. The conversion of natural forests and grasslands to agriculture and livestock is the biggest cause of global biodiversity loss. The next biggest drivers are the exploitation of wildlife through fishing, logging, trade and hunting – and then pollution. Climate change ranks fourth on land so far but second in oceans. This is the main result of an international study led by researchers from Universidad Nacional de Córdoba (UNC) in Argentina, Helmholtz Centre for Environmental Research (UFZ), German Centre for Integrative Biodiversity Research (iDiv) and the Natural History Museum London. The study, published in Science Advances, demonstrates that fighting climate change alone will not be enough to prevent the further loss of biodiversity.
More10.11.2022 | Biodiversity Conservation, Media Release, TOP NEWS
Protecting and connecting nature across Europe
Based on a media release by the International Institute for Applied Systems Analysis (IIASA) Vienna/Leipzig. Europe needs healthy ecosystems that benefit biodiversity and people and are resilient to climate change. The Horizon Europe NaturaConnect Project will support European Union governments and other public and private institutions in designing a coherent, resilient and well-connect Trans-European Nature Network. The new project is conducted by international partners from research and environmental organisations, led by the International Institute for Applied Systems Analysis (IIASA), the German Centre for Integrative Biodiversity Research (iDiv) and the Martin-Luther University Halle-Wittenberg (MLU).
More24.10.2022 | Biodiversity Synthesis, Media Release, Physiological Diversity, TOP NEWS
More yield, fewer species: How human nutrient inputs alter grasslands
Leipzig. With high nutrient inputs in grasslands, more plant species get lost over longer periods of time than new ones can establish. In addition, fewer new species settle than under natural nutrient availability. With a worldwide experiment, researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ) and the Martin Luther University Halle-Wittenberg (MLU) have now been able to show why additional nutrient inputs reduce plant diversity in grasslands. Another finding was that the increase in biomass with nutrient inputs is due to a few plant species that can use higher nutrient inputs to their advantage and remain successfully at a site over long periods of time. The results have been published in the journal Ecology Letters.
More20.10.2022 | iDiv Members, Media Release, TOP NEWS
Smartphone data can help create global vegetation maps
Leipzig. Missing knowledge in the global distribution of plant traits could be filled with data from species identification apps. Researchers from Leipzig University, the German Centre for Integrative Biodiversity Research (iDiv) and other institutions were able to demonstrate this based on data from the popular iNaturalist app. Supplemented with data on plant traits, iNaturalist input results in considerably more precise maps than previous approaches based on extrapolation from limited databases. Among other things, the new maps provide an improved basis for understanding plant-environment interactions and for Earth system modelling. The study has been published in the journal Nature Ecology and Evolution.
More19.10.2022 | iDiv Members, Media Release, TOP NEWS
Ecological imbalance: How plant diversity in Germany has changed in the past century
Based on a media release by Martin Luther University Halle-Wittenberg (MLU) Halle. Germany’s plant world has seen a greater number of losers than winners over the past one hundred years. While the frequencies and abundances of many species have shrunk, they have significantly increased in others. This has resulted in a very uneven distribution of gains and losses. It indicates an overall, large-scale loss of biodiversity, as a team led by the Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) reports in Nature.
More17.10.2022 | iDiv Members, Media Release, TOP NEWS
European colonial legacy is still visible in today’s alien floras
Based on a media release by the University of Vienna Vienna/Leipzig. Alien floras in regions that were once occupied by the same European power are, on average, more similar to each other compared to outside regions and this similarity increases with the length of time a region was occupied. This is the conclusion of a study by an international team of researchers led by the University of Vienna and with the participation of researchers from the German Centre for Integrative Biodiversity Research (iDiv). The results were recently published in the scientific journal Nature Ecology and Evolution.
More12.10.2022 | Experimental Interaction Ecology, Media Release, TOP NEWS
Global study: Few of the ecologically most valuable soils are protected
Halle, Leipzig, Seville. Current protected areas only poorly cover the places most relevant for conserving soil ecological values. This is the conclusion of a new study published in the journal Nature. To assess global hotspots for preserving soil ecological values, an international team of scientists measured different dimensions of soil biodiversity (local species richness and uniqueness) and ecosystem services (like water regulation or carbon storage). They found that these dimensions peaked in contrasting regions of the world. For instance, temperate ecosystems showed higher local soil biodiversity (species richness), while colder ecosystems were identified as hotspots of soil ecosystem services. In addition, the results suggest that tropical and arid ecosystems hold the most unique communities of soil organisms. Soil ecological values are often overlooked in nature conservation management and policy decisions; the new study, published in Nature, demonstrates where efforts to protect them are needed most.
More01.10.2022 | Macroecology and Society, Media Release, TOP NEWS
Land tenure drives deforestation rates in Brazil
Leipzig. Tropical deforestation causes widespread degradation of biodiversity and carbon stocks. Researchers from the German Center of Integrative Biodiversity Research (iDiv) and Leipzig University were now able to test the relationship between land tenure and deforestation rates in Brazil. Their research, which was published in Nature Communications, shows that poorly defined land rights go hand in hand with increased deforestation rates. Privatising these lands, as is often promoted in the tropics, can only mitigate this effect if combined with strict environmental policies.
More29.09.2022 | Biodiversity Conservation, Media Release, Research, TOP NEWS
Historical reduction of the wolf in the Iberian Peninsula
Based on a media release of the Consejo Superior de Investigaciones Científicas, Spain (CSIC) Seville/Leipzig. The distribution of the wolf covered at least 65% of the Iberian Peninsula in the mid-19th century. Compared to this finding, recent expansions mean little more than a stabilisation of the species. This is the result of a study led by the Doñana Biological Station of the Consejo Superior de Investigaciones Científicas (EBD-CSIC) in collaboration with the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU). To reach these conclusions, the research team analysed the historical information from a geographical survey collected in the mid-19th century. The study has been published in the journal Animal Conservation. It provides new information on the history of wolf declines due to human persecution.
More07.09.2022 | Media Release, sDiv, TOP NEWS
Location, location, location! What leads to lignification on islands
Based on a media release from Philipps-Universität Marburg. Marburg/Leipzig/Leiden. Increased drought, the lack of predators and isolation lead to a tendency of plants on islands to become woody. The location of the islands on which the species concerned are native also plays a role. This is the result of a study led by the Philipps University of Marburg and the German Centre for Integrative Biodiversity Research (iDiv) together with the Naturalis Biodiversity Centre in Leiden and other institutions. The study by the German-Dutch research team, which has now been published in the scientific journal PNAS, shows how islands act as natural laboratories of evolution.
More01.09.2022 | iDiv Members, Media Release, Research, TOP NEWS
High plant diversity is often found in the smallest of areas
Based on a media release by Martin Luther University Halle-Wittenberg Halle/Leipzig. The steppes of Eastern Europe are home to a similar number of plant species as the regions of the Amazon rainforest. However, this is only apparent when species are counted in small sampling areas rather than hectares of land. An international team of researchers led by the Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig has now shown how much estimates of plant diversity change when the sampling area ranges from a few square metres to hectares. Their results have been published in the journal Nature Communications and could be utilised in new, more tailored nature conservation concepts.
More15.08.2022 | Evolutionary Ecology, Media Release, Science-Policy, Science-Policy, TOP NEWS
National parks – islands in a desert?
Leipzig/Jena/Bonn. How effective is biodiversity conservation of European and African national parks? This seems to be strongly associated with societal and economic conditions. But even under the most favourable conditions, conservation efforts cannot completely halt emerging threats to biodiversity if conditions outside of the parks do not improve. This is the conclusion of a new study led by the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology (MPI EVA), and the University of Bonn, in collaboration with the Helmholtz Centre for Environmental Research (UFZ), Leipzig University, the Friedrich Schiller University Jena and many other institutions. The study published in the journal Nature Sustainability highlights the urgent need for a better design of national park networks.
More27.07.2022 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS
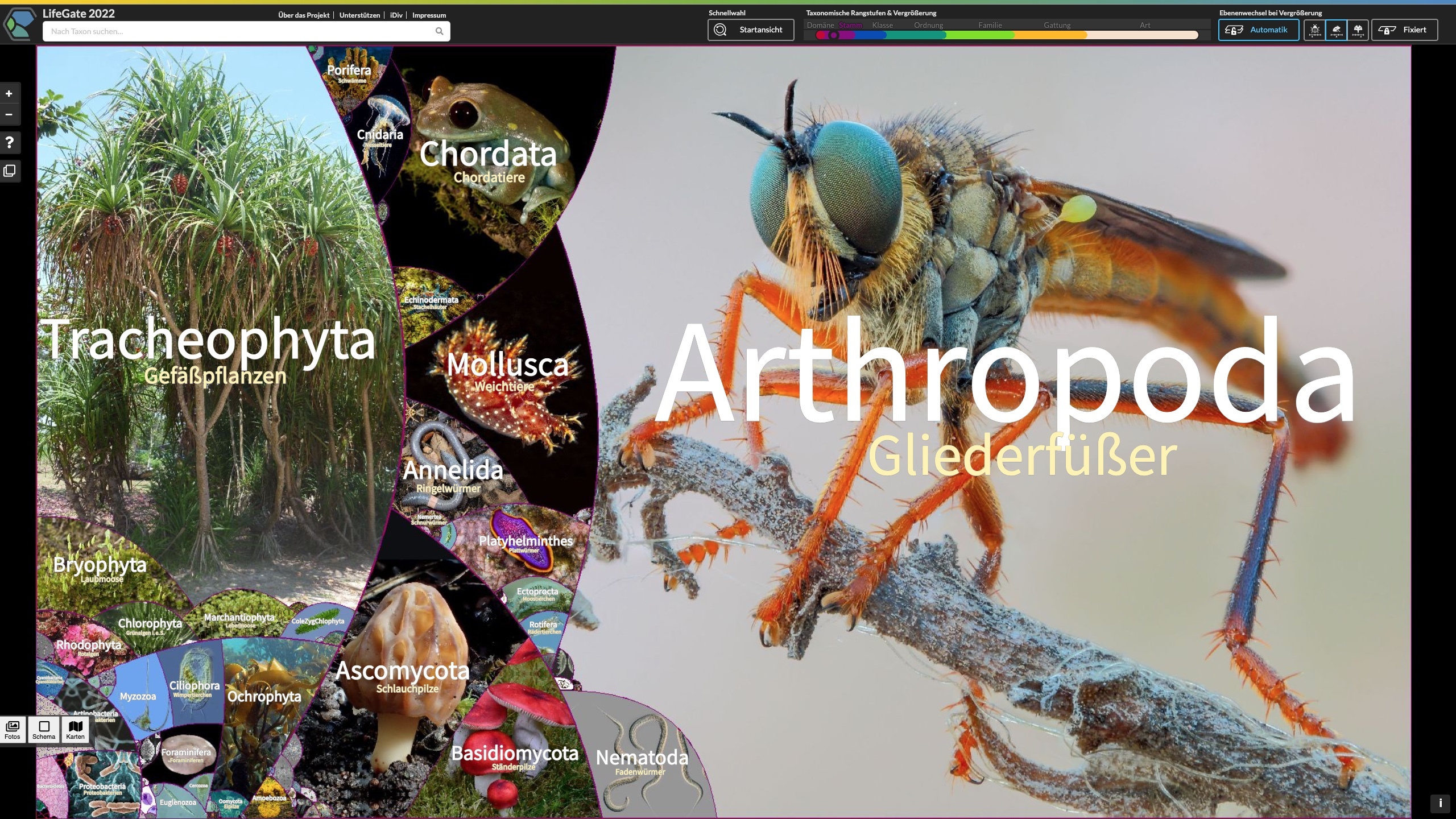
LifeGate – New interactive map shows the full diversity of life
Joint press release of Leipzig University (Botanical Garden) and the German Centre for Integrative Biodiversity Research (iDiv) Leipzig. Researchers from Leipzig have published a gigantic digital map displaying the full diversity of life through thousands of photos. The so-called LifeGate encompasses all 2.6 million known species of this planet and shows their relationship to each other. The interactive map can now be accessed free of charge at lifegate.idiv.de.
More19.07.2022 | Biodiversity Conservation, Media Release, TOP NEWS
Informing future conservation priorities of ecosystems in the Tropical Andes
Joint press release by iDiv and NatureServe Washington D.C./Leipzig. Only about 5% of ecosystems within the Tropical Andes biodiversity hotspot are adequately represented in designated protected areas. Representation may be determined by a series of targets proposed by the Convention on Biological Diversity (CBD), which states at least 30% of land and waters in each country should be conserved. Protecting the full diversity of ecosystems reduces extinction risk for the species those ecosystems support, but very few places on the planet currently meet the CBD target. There is an opportunity to increase representation of ecosystems that meet the CBD target to 31% (39 ecosystem types total) across four Andean countries (Bolivia, Colombia, Ecuador, and Peru) through the additional protection by governments and civil societies of Key Biodiversity Areas (KBAs), places that meet the internationally recognized standard for sites that significantly contribute to the global persistence of biodiversity. This conclusion is shown in a new study led by NatureServe, the German Centre for Integrative Biodiversity Research (iDiv), and the Martin Luther University Halle-Wittenberg (MLU), together with other institutions throughout the U.S., Europe, and South America. The study, now published in the journal Remote Sensing, demonstrates how Essential Biodiversity Variables (EBVs) coupled with KBAs can inform conservation decisions at multiple scales.
More18.07.2022 | Experimental Interaction Ecology, Media Release
More species could be threatened with extinction than previously thought
Based on a media release by the University of Minnesota Saint Paul/Leipzig. On average, 30 per cent of all species worldwide might be threatened with extinction or have already become extinct over the last 500 years. This was the result of estimates by 3,331 experts working on biodiversity in 187 countries. This large and diverse group of experts was asked in a survey, led by researchers from the University of Minnesota and with the participation of iDiv and the University of Leipzig, to provide assessments of the change in the species they study. The results are expected to reduce knowledge gaps in existing scientific assessments of global biodiversity and thus improve the knowledge base for policy decisions. The study was published in the journal Frontiers in Ecology and the Environment.
More14.07.2022 | Biodiversity Synthesis, Media Release, TOP NEWS
North American birds not fully adjusting to changing climate
Leipzig/Halle/Seville. Some species of birds in North America have not fully adjusted their distributions in response to ongoing climate change. The places these birds live have become more decoupled from their optimal climate conditions, while other features of the environment become more constraining. This trend of climate decoupling is more pronounced for habitat specialists and for species declining in abundance. These are the results of a study by the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University, the Martin Luther University Halle-Wittenberg (MLU) and the Doñana Biological Station, now published in Nature Ecology & Evolution. Climate decoupling as a result of ongoing climate change may lead to additional stresses on many species of birds, and exacerbate population declines.
More08.07.2022 | Biodiversity and People, Media Release, TOP NEWS
Protecting butterflies in Leipzig
Based on a media release by Helmholtz Centre for Environmental Research (UFZ) The Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv), the Office for Urban Green Spaces and Waters of the City of Leipzig, and BUND Leipzig are now in their third season with the citizen science project “VielFalterGarten” this summer. The aim of the project, which is part of the Bundesprogramm für Biologische Vielfalt, is to research and protect the butterfly diversity in the city of Leipzig together with citizens.
More30.06.2022 | Biodiversity and People, Media Release, TOP NEWS
Six guiding principles for the EU agricultural policy to halt biodiversity loss
Leipzig/Rostock. To halt biodiversity loss, the future design of EU agricultural policy could be guided by six basic principles and accompanied by multi-annual agreements and progressive payment systems. These are at the core of recommendations made by over 300 scientists from 23 EU member states who were consulted at the request of the European Commission. The process was coordinated by the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ), Thünen Institute of Rural Studies, and the University of Rostock. A synthesis of the results of this extensive consultation process has now been published in the journal Conservation Letters.
More29.06.2022 | iDiv Members, Media Release, TOP NEWS
Life in the earth’s interior as productive as in some ocean waters
Joint media release by iDiv and Friedrich Schiller University Jena Jena. Microorganisms in aquifers deep below the earth’s surface produce similar amounts of biomass as those in some marine waters. This is the finding of researchers led by the Friedrich Schiller University Jena and the German Centre for Integrative Biodiversity Research (iDiv). Applying a unique, ultra-sensitive measurement method using radioactive carbon, they were able to demonstrate for the first time that these biotic communities in absolute darkness do not depend on sunlight. Instead, they can obtain energy autonomously from rock oxidation or from compounds transported into the deep. The study has been published in Nature Geoscience.
More24.06.2022 | iDiv, Media Release, TOP NEWS
Junior researchers investigate the impact of human activities on soil
Joint media release by Leipzig University and iDiv Leipzig. “Mysterious but beautiful” – this is how Dr Anton Potapov describes the world of organisms that live under our feet. Starting on 1 July 2022, the ecologist and microbiologist is setting up a new junior research group at the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University. It will take a closer look at the role of soil organisms in the ecosystem. Potapov’s work during the coming years will receive 1 million euros in funding through the German Research Foundation’s (DFG) Emmy Noether Programme.
More15.06.2022 | Biodiversity Conservation, TOP NEWS
The Gollum effect and its impacts on the scientific community
Report by Dr Jose Valdez, Postdoctoral Researcher of the Biodiversity Conservation research group at iDiv and Leipzig University, and senior author of a new publication in Frontiers in Ecology and Evolution While scientific research is becoming increasingly more collaborative, the ability to freely conduct research remains restricted by established researchers who feel they have the sole right to study specific study sites, model organisms, research topics, and sometimes even entire scientific fields. Like Gollum from The Lord of the Rings, they become possessively attached and guard these aspects of their research. In our recent publication in Frontiers in Ecology and Evolution we refer to this as the ‘Gollum effect’ and explore ways in which this can be overcome to promote scientific openness and lead science into a new era where it is dispensed fairly instead of at the hands of a selected few.
More09.06.2022 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
What doesn’t kill you makes you stronger
Based on a media release by the University of Zurich Zurich, Leipzig. The exposure to drought during previous generations in the field increases complementarity between offspring of different grassland species and thus makes them more resilient to subsequent drought. An international research team led by the University of Zurich and with the participation of the German Centre for integrative Biodiversity research and Leipzig University has revealed this transgenerational effect with about 1000 experimental plant communities in pots. The results, published in Nature Communications, suggest that if past extreme climatic events do not completely exclude species, they may enhance the sustainability of biodiversity and ecosystem functioning in a future with more frequent extreme events.
More06.06.2022 | iDiv, Media Release, Symbiont Evolution, TOP NEWS
Evolution: New Emmy Noether group investigates interactions between bacteria and insects
Joint press release by Martin Luther University Halle-Wittenberg (MLU) and iDiv Halle/Leipzig. The German Research Foundation (DFG) has accepted the biologist Dr Michael Gerth into the Emmy Noether Programme and will fund his work with up to 1.4 million euros in the coming years. With this funding, Gerth will move from the UK to the Martin Luther University Halle-Wittenberg (MLU) and establish his own working group at the German Centre for Integrative Biodiversity Research (iDiv). His research focuses on the evolution of special bacteria, which live in flies, bees and other insects.
More03.06.2022 | Biodiversity and People, Media Release, TOP NEWS
Heat-lovers are the lucky ones
Based on a media release from the Technical University of Munich (TUM) Munich/Leipzig. Heat-loving insect species in Germany are benefiting from global warming. The trends of different insect groups vary considerably. While the populations of butterflies and grasshoppers decreased more frequently than they increased, dragonflies showed predominantly positive trends. This is the conclusion of a new study led by the Technical University of Munich (TUM), the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich Schiller University Jena. In order to close significant data gaps, the research team evaluated an extensive collection of previously little-used data on over 200 insect species in Bavaria since 1980. The study was published in Global Change Biology.
More02.06.2022 | Evolutionary Ecology, Media Release, TOP NEWS
The largest genomic catalogue of wild chimpanzees
Based on a media release by the Institute of Evolutionary Biology (IBE) Barcelona/Leipzig. An international research team led by the Institute of Evolutionary Biology (IBE), the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology (MPI EVA) and Leipzig University has compiled the largest genomic catalogue of wild chimpanzee populations in Africa. For the first time, genomic information, obtained from hundreds of faecal samples of chimpanzees from across their range, has been sequenced, clarifying their evolutionary history and contributing to the conservation of these great apes. The new genomic atlas, published in the journal Cell Genomics, has the potential to map routes and sources of illegal trafficking that could be used to protect this endangered species.
More01.06.2022 | Media Release, Theory in Biodiversity Science, TOP NEWS
Photos of Amazon animals supply extensive collection of biodiversity data
Manaus/São Paulo/Jena/Leipzig. An international team of researchers has published the largest collection of data from camera traps on Amazonian rainforest animals. The collection currently encompasses over 120,000 registers, plus information on time and location. It will improve research on the abundance, diversity and habitat conditions of jaguars, toucans, harpy eagles and many other endangered rainforest species and contribute to their protection. 147 scientists from 122 research institutions and nature conservation organisations collaborated under the leadership of the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich Schiller University of Jena to build this new database, which is based on camera trap photos. It has now been published in the journal Ecology.
More31.05.2022 | iDiv Members, Media Release, TOP NEWS
Healthy development thanks to older siblings
Based on a media release by Max Planck Institute for Evolutionary Anthropology (MPI-EVA) Leipzig. Even mild forms of stress in pregnant women can have negative effects on their children’s behavior even years after birth. Older siblings, however, can mitigate this effect. This is what researchers from the Helmholtz Centre for Environmental Research (UFZ), Leipzig University (UL), the Max Planck Institute for Evolutionary Anthropology (MPI-EVA) and the German Center for Integrative Biodiversity Research (iDiv) have found. The study was published in the journal BMC Public Health.
More18.05.2022 | Biodiversity Conservation, Media Release, TOP NEWS
Native plant gardening for species conservation
Halle/Leipzig. Declining native species could be planted in urban green spaces. Researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU), Leipzig University and other institutions describe how to use this great potential for species protection. In their most recent study, published in the journal Nature Sustainability, they recommend practical conservation gardening methods in a bid to restructure the horticultural industry and reverse plant species declines.
More17.05.2022 | TOP NEWS
iDiv honours Professor Beate A. Schücking
Leipzig. iDiv has honoured Professor Beate A. Schücking’s many years of commitment to the German Centre for Integrative Biodiversity Research (iDiv). During a ceremony on Monday, iDiv presented her with a tree sponsorship in the Leipzig University Botanical Garden. As Rector of the university and as a member of the iDiv Board of Trustees, Professor Schücking has played a major role in the success story of the research centre over the past eleven years.
More11.05.2022 | Experimental Interaction Ecology, Research, TOP NEWS
Earthworms increase the stability of soil organic carbon
Report by Dr Gerrit Angst, postdoctoral researcher of the Experimental Interaction Ecology at iDiv and Leipzig University and first author: Leipzig/Budweis/Munich. Earthworm activity stimulates microorganisms to accelerate the production of more stabilised soil organic carbon. This is the conclusion from a synthesis of recent literature collated by a team led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University, the Czech Academy of Sciences, and the Technical University of Munich. Based on this synthesis, the authors developed a novel concept of the role of earthworms in soil organic carbon dynamics that aids management of soils as a carbon sink. The study has recently been published in Global Change Biology.
More09.05.2022 | iDiv, iDiv Members, Media Release, TOP NEWS
More than four million euros for research on trees and shrubs
Halle/Leipzig/Jena/Beijing. The International Research Training Group TreeDì can continue its work. On Friday, the relevant Grants Committee of the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) approved its continued funding with more than four million euros until 2027. The programme is conducted in cooperation with the Martin Luther University Halle-Wittenberg (MLU), the Friedrich Schiller University Jena and Leipzig University as well as the University of the Chinese Academy of Sciences (UCAS). The aim of the research is to better understand the interactions of trees and shrubs in forests. On the German side, the doctoral researchers work and interact at the German Centre for Integrative Biodiversity Research (iDiv), and a large number of iDiv scientists are involved in the TreeDì research and qualification programme, for example, in teaching.
More29.04.2022 | Evolution and Adaptation, Media Release, TOP NEWS
Dinosaur extinction changed plant evolution
Leipzig, Amsterdam, Zurich. The absence of large herbivores after the extinction of the dinosaurs changed the evolution of plants. The 25 million years of large herbivore absence slowed down the evolution of new plant species. Defensive features such as spines regressed and fruit sizes increased. The research led by the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University has demonstrated this using palm trees as a model system. The researchers were also able to show how profound the changes were: Even the reappearance of large herbivores millions of years later could only partially overwrite the changes that had already taken place. The study has been published in the journal Proceedings of the Royal Society B. It provides a view of the geological past and, at the same time, promotes a better understanding of the consequences of current extinction processes.
More27.04.2022 | Biodiversity Conservation, Media Release, TOP NEWS
More than one in five reptile species are threatened with extinction
Based on a media release by NatureServe, the International Union for Conservation of Nature (IUCN), and Conservation International Virginia, Halle. At least 21% of all reptile species globally are threatened with extinction, according to a new study led by NatureServe, the International Union for Conservation of Nature (IUCN), and Conservation International, with contributions from the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU). The study, published in the journal Nature, also shows that conservation efforts for other animals also contribute to the preservation of many reptile species.
More26.04.2022 | Biodiversity and People, Media Release, TOP NEWS, UFZ-News
Generating knowledge together – The Citizen Science Strategy 2030
Based on a media release by the Helmholtz Centre for Environmental Research (UFZ) Berlin/Leipzig. Generating knowledge together – that is the goal of Citizen Science. On 29 April 2022, scientists from the Helmholtz and Leibniz Associations as well as the German Centre for Integrative Biodiversity Research (iDiv) will present the Citizen Science Strategy 2030 for Germany to the public and present a series of current projects at a Citizen Science Festival. The strategy addresses the greatest challenges and potentials of Citizen Science over the next ten years. It contains 94 specific recommendations for action to further develop citizen science in Germany and to integrate it permanently into science, society and politics. The Citizen Science Strategy 2030 Germany was developed in a two-year participatory process with more than 200 actors from 136 organisations. The process was largely funded by the Deutsche Bundesstiftung Umwelt (DBU) and the Federal Ministry of Education and Research (BMBF).
More07.04.2022 | iDiv, iDiv Members, Media Release, TOP NEWS
Christian Wirth receives Leipzig Science and Humanities Prize 2022
Based on a media release by the City of Leipzig, Leipzig University and the Saxon Academy of Sciences and Humanities in Leipzig Leipzig. This year’s Leipzig Science and Humanities Prize goes to Christian Wirth, who is a professor for special botany and functional biodiversity at Leipzig University, a Fellow at the Max Planck Institute for Biogeochemistry in Jena, and Speaker of the German Centre for Integrative Biodiversity Research (iDiv). With this prize, the City of Leipzig, Leipzig University and the Saxon Academy of Sciences and Humanities in Leipzig honour researchers who meet the highest academic standards and thus reinforce Leipzig’s reputation as a city of science with a national and international reputation for its research and education landscape.
More31.03.2022 | Biodiversity Conservation, iDiv Members, TOP NEWS
How can we improve biodiversity monitoring in Europe?
Based on a press release by the Martin Luther University Halle-Wittenberg (MLU) National biodiversity monitoring programmes in Europe face many challenges: too little coordination, inadequate technical and financial resources as well as unclear targets. This is the conclusion of an initial policy report by the Europe-wide project “EuropaBON”. The analysis includes data from more than 350 experts in policy, science and environmental protection. The team is also drafting a proposal for the transnational monitoring of Europe’s biodiversity and ecosystems.
More30.03.2022 | Experimental Interaction Ecology, Media Release, TOP NEWS
European earthworms reduce insect populations in North American forests
Leipzig/Calgary. Earthworms introduced into northern North America have a negative impact on the insect fauna above ground. Soil ecologists, led by the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University describe this observation in the journal Biology Letters. The researchers found this impact for abundance as well as for biomass and species richness of insects. Their results indicate that changes in insect communities can have causes that have previously received little attention. These should be given greater consideration in nature conservation.
More11.03.2022 | Biodiversity and People, TOP NEWS
Citizens, conservation and climate change
Biologists and botanists from the universities of Berlin, Halle, Jena and Leipzig, as well as the German Centre for Integrative Biodiversity Research (iDiv) and the Helmholtz Centre for Environmental Research (UFZ) invite citizens to participate in scientific research. In the “Plant ClimateCulture!” project, they can help to record the effects of climate change in cities by observing plants in their own and in the botanical gardens of the participating cities. The project results also contribute to making cities more climate-adapted and thus more sustainable and attractive to live in. Insights into the project and a lot of background information will be provided during a tour of the Botanical Garden of the University of Leipzig (Linnéstraße 1) on 20 March.
More23.02.2022 | Biodiversity Synthesis, Media Release, TOP NEWS
While some insects are declining, others might be thriving
Leipzig/Halle/Jena. Observations of abundance changes in one group of insects– for example grasshoppers – say very little about how other types of insects, such as flies, are doing, even in the same place. This is because different groups of insects may show similar trends in one place, but dissimilar trends in other places. These are the findings of a new meta-study systematically examining long-term data on insects from more than 900 locations worldwide. The study, published in Biology Letters, was led by a team of researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU), the Friedrich Schiller University Jena and the Helmholtz Centre for Environmental Research (UFZ). It highlights the importance of monitoring multiple groups of species simultaneously to provide guidelines for conservation policies.
More07.02.2022 | Biodiversity Conservation, Media Release, TOP NEWS
7 to 9 percent of all European vascular plants are globally threatened
Halle, Leipzig. Seven to nine percent of all vascular plant species occurring in Europe are globally threatened. This is the result of a study led by the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg and Leipzig University. The researchers combined Red Lists of endangered plant species in Europe with data on their global distribution. The study has been published in the journal Plants, People, Planet. It helps assess the overall level of threat to plant species and thus supports the basis of international nature conservation activities.
More03.02.2022 | Biodiversity and People, Media Release, TOP NEWS
Love of nature is partially heritable, study of twins shows
Based on a press release by PLoS Biology Singapore/Exeter/Brisbane/Leipzig. A person’s appreciation of nature and their tendency to visit natural spaces are heritable characteristics. This is the result of a large-scale study of UK twins between the National University of Singapore, the University of Exeter, the University of Queensland, the Helmholtz Centre for Environmental Research (UFZ), and the German Centre for Integrative Biodiversity Research (iDiv). The study has now been published in the journal PLoS Biology.
More18.01.2022 | Media Release, Molecular Interaction Ecology, TOP NEWS
Trees call for help from birds and predatory insects
Leipzig/Jena. Trees emit scents when attacked by caterpillars and other herbivores. They use these to attract predatory insects and even birds, thus getting rid of their pests. This had only been demonstrated in smaller-scale experiments so far. A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Friedrich Schiller University Jena and Leipzig University has now demonstrated this phenomenon for the first time in a natural habitat – in the 40-metre-high canopy of the Leipzig floodplain forest. The chemical calls for help are so effective that they significantly determine the composition of the insect community in the canopy. This knowledge could be used for future natural pest control in agriculture and forestry, the researchers write in the journal Ecology Letters.
More17.01.2022 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS, UFZ-News
Can “rewilding” synergize biodiversity conservation and sustainable regional development?
Based on a media release by Rewilding Oder Delta (ROD) and the UFZ The research project “REWILD_DE – Conservation of biodiversity and valorisation of ecosystem services through rewilding – learning from the Oder Delta”, funded by the German Federal Ministry of Education and Research (BMBF), started in November 2021. Scientists and practitioners are working together to explore the potential of rewilding for the restoration of biodiversity and the promotion of nature-based economic development in the model area of the Oder Delta in western Pomerania (north-eastern Germany). Furthermore, they will investigate to what extent the experiences and results gained during the project can be transferred beyond the project area and serve as a model for other regions. The project is coordinated at the Helmholtz Centre for Environmental Research (UFZ) with project 3 partners: the German Centre for Integrative Biodiversity Research (iDiv) at the Martin Luther University Halle-Wittenberg (MLU), the Eberswalde University for Sustainable Development (HNEE) and the association Rewilding Oder Delta (ROD), a practice partner in the study area.
More14.01.2022 | iDiv, Media Release, TOP NEWS
Implementing global biodiversity targets in Central Germany
Leipzig. The environment ministers of the Central German states and representatives from nature conservation organizations and authorities today exchanged views on the implementation of the international biodiversity targets at the state level. The virtual meeting took place at the initiative of the German Center for Integrative Biodiversity Research (iDiv) in Leipzig and is part of a long-term dialogue.
More04.01.2022 | Biodiversity and People, TOP NEWS
How to use ecosystem services in social-ecological networks
Leipzig. Social-ecological networks (SEN) have recently been proposed as a promising approach to conceptualise and analyse interactions and outcomes in social-ecological systems. In a social system, networks represent interactions between actors, such as knowledge exchange, trust, collaboration, which influence governance effectiveness. In ecology, networks depict biological nodes (e.g. species, patches) and processes that connect them (e.g., predation, landscape connectivity). Despite similarities in the approaches applied in both social sciences and ecology, these areas have developed largely in parallel without capitalising on their complementarity. As a result, there is limited integration of social and ecological theories, inclusion of ecological complexity in governance research, and vice versa. Building from different disciplines, in our new article in Trends in Ecology & Evolution we propose a typology to represent ecosystem service in SENs and identify opportunities and challenges of using SENs in ecosystem service research.
More23.12.2021 | iDiv Members, Media Release, TOP NEWS
Climate and soil determine the distribution of plant traits
Based on a press release by the Max Planck Institute for Biogeochemistry Zurich/Jena/Leipzig. An international research team succeeded in identifying global factors that explain the diversity of form and function in plants. A team led by researchers from the University of Zurich, the Max Planck Institute for Biogeochemistry, Leipzig University and the German Centre for Integrative Biodiversity Research (iDiv), collected and analysed plant data from around the world. Their publication in Nature Ecology and Evolution for the first time shows for characteristics such as plant size, structure, and life span, how strongly these are determined by climate and soil properties. Insights derived from this could be crucial to improving Earth system models with regard to the role of plant diversity.
More20.12.2021 | iDiv Members, Media Release, TOP NEWS
Extreme droughts also afflicting the Leipzig floodplain forest
Leipzig. The Leipzig floodplain forest was not equipped to endure two consecutive hotter drought years. Although the trees were able to partially cope with the 2018 drought, the accumulated and ongoing damage from drought stress caused their growth to collapse in the second drought year 2019. Depending on the tree species, this decrease in growth ranged from 9% to 42% when compared to climatically normal years. This is the result of a study published by a team led by the German Centre for Integrative Biodiversity Research (iDiv) in the journal Global Change Biology. The study may enable better understanding and prediction of how forests respond to climate change.
More17.12.2021 | iDiv Members, Media Release, TOP NEWS
Well insured: Forests with many tree species grow more consistently
Leipzig / Beijing. The annual growth of forests fluctuates due to extreme climate conditions such as drought and heavy rainfall. These fluctuations are less pronounced in species-rich forests than in those with low species richness. Furthermore, forests with many different tree species produce more wood. It is not only a question of the species’ richness, but also the diversity of their functional characteristics. This has been shown by an international research team led by the German Centre for Integrative Biodiversity Research (iDiv) and the Chinese Academy of Sciences Beijing (CAS). The results of the study have been published in the journal Science Advances. They provide significant impetus for ways in which forestry can promote more reliable productivity.
More13.12.2021 | iDiv, iDiv Members, Media Release, TOP NEWS
Sensational discovery in the Leipzig floodplain forest
Based on a media release by the Leipzig University Leipzig. Researchers and biology students from the University of Leipzig have discovered an extremely rare insect during a zoological field trip in the Leipzig floodplain forest: Bittacus hageni, belonging to the order of scorpionflies. The researchers caught several specimens in the leaf canopy of the Leipzig floodplain forest. This species is only found in very few places in Europe. In Germany, it was considered extinct until 2003. The find, which the researchers describe in the journal Entomologische Nachrichten und Berichte, highlights the importance of the Leipzig floodplain forest as a unique but endangered habitat. But it also proves the importance of student excursions to make valuable discoveries possible.
More12.12.2021 | Research, sDiv, TOP NEWS
Uniqueness is disappearing
Based on a press release by the University of Konstanz Konstanz/Leipzig. Plant communities all over the planet are becoming more and more alike, even between regions far removed from each other. The reason is the dispersal of non-native plant species, according to the result of a global research project led by biologists from Konstanz and with the participation of the German Centre for Integrative Biodiversity Research (iDiv). The results of the study, which were published in the journal Nature Communications, highlight the importance of effective protective measures in order to maintain regional uniqueness.
More10.12.2021 | Biodiversity and People, Media Release, TOP NEWS
Enjoying nature conservation and doing something good for nature
Based on a media release by the Thünen Institute of Biodiversity
More09.12.2021 | Biodiversity Economics, Media Release, sDiv, TOP NEWS
Let tropical forests grow again
Based on a media release by Wageningen University Wageningen/Leipzig. Tropical forests are vanishing at an alarming rate through deforestation, but also have the potential to regrow naturally on abandoned lands. A study published this week in Science shows that regrowing tropical forests recover surprisingly fast. After 20 years, characteristic attributes have recovered by an average of nearly 80% of old-growth forest values. This applies e.g. to soil fertility, soil carbon storage, tree diversity, plant functioning and forest structure. The study led by Wageningen University with support from the German Centre for Integrative Biodiversity Research (iDiv) concludes that natural regeneration is a low-cost, nature-based solution for climate change mitigation, biodiversity conservation, and ecosystem restoration.
More09.12.2021 | Biodiversity Conservation, Media Release, sDiv, TOP NEWS
Widespread plants displace rarer species across habitats
Based on a press release by the Austrian Academy of Sciences (ÖAW) Leipzig/Vienna. Widespread species are on the rise in several European ecosystems and are replacing rarer plant species. As a result, distinct habitats are becoming increasingly similar in species composition. An international team led by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) with the participation of the Austrian Academy of Sciences (ÖAW) has found supporting evidence for this development in mountain summits as well as in forests and lowland grasslands. A proposed reason for this species shift could be increased nutrient levels in soils caused by agriculture and air pollution. The results of the study have been published in the journal Ecology Letters.
More01.12.2021 | Biodiversity Economics, Media Release, TOP NEWS
Climatic changes and overfishing depleted Baltic herring long before industrialisation
Joint media release with Kiel University Kiel/Leipzig. The collapse of the important herring fishery in the western Baltic Sea towards the end of the 16th century was the result of a combination of overfishing and climate change. The researchers see similarities in the current development of German fish stocks. A team of fisheries economists, historians and biologists from Kiel University (CAU), the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University has reconstructed herring catch levels in the Baltic Sea between 1200 and 1650. The by far most important fishery for autumn spawning herring at that time collapsed within a very short period, without recovering until today. The study, for which they evaluated historical sources, was published in the journal Hansische Geschichtsblätter.
More23.11.2021 | Biodiversity and People, Biodiversity Conservation, iDiv, iDiv Members, Media Release, TOP NEWS
From ambition to biodiversity action: Time to hold actors accountable
Leipzig. To achieve global goals for biodiversity conservation, the national-level implementation must be significantly improved. National policy instruments need to precisely define effective actions and the actors responsible for implementation. Accountability needs to be ensured through systematic monitoring of progress. These recommendations are at the core of a 3-step framework proposed by an international team of scientists led by the German Centre for Integrative Biodiversity Research (iDiv), published in the journal Conservation Letters. The authors stress the need for urgency to avoid repeating failures of past international agreements and to move to effective implementation of agreed policy targets. One mistake, in particular, should be avoided.
More16.11.2021 | TOP NEWS
Highly Cited Researchers 2021
Clarivate Analytics lists eight iDiv members in its 2021 selection of “Highly Cited Researchers”. According to Clarivate Analytics, these scientists have demonstrated significant influence through publication of multiple papers, highly cited by their peers, during the last decade.
More03.11.2021 | Media Release, Research, Theory in Biodiversity Science, TOP NEWS
Where the wild bears are
Leipzig/Jena/Oxford. New software helps determine the movements of large wild animals thereby minimising conflicts with people. The software is simpler than measurements obtained using radio transmitters and can be used where conventional methods fail. An international team led by the German Centre for Integrative Biodiversity Research (iDiv), the Friedrich Schiller University Jena, Aarhus University and the University of Oxford has published a description of the new software in the journal Methods in Ecology and Evolution.
More14.10.2021 | Media Release, Species Interaction Ecology, TOP NEWS
Decline of plant pollinators threatens biodiversity
Joint media release with University of Konstanz, based on a media release by Stellenbosch University, South Africa Stellenbosch, Konstanz, Leipzig. About 175 000 plant species – half of all flowering plants – mostly or completely rely on animal pollinators to make seeds and so to reproduce. This is the finding of a study conducted by a global research network with participation of the German Centre for Integrative Biodiversity Research (iDiv) and the University of Konstanz, recently published in the journal Science Advances. Declines in pollinators could therefore cause major disruptions in natural ecosystems, including loss of biodiversity.
More13.10.2021 | Evolution and Adaptation, Media Release, TOP NEWS
Geologically vibrant continents produce higher biodiversity
Based on a media release from ETH Zurich Leipzig, Zurich. Using a new mechanistic model of evolution on Earth, researchers at German Centre for Integrative Biodiversity Research (iDiv) and ETH Zurich can now better explain why the rainforests of Africa are home to fewer species than the tropical forests of South America and Southeast Asia. The key to high species diversity lies in how dynamically the continents have evolved over time.
More06.10.2021 | Biodiversity Conservation, Media Release, TOP NEWS
More and more roads but little knowledge about their impact on wildlife
Based on a media release by the Complutense University of Madrid (UCM) Madrid/Leipzig/Halle. In regions with a very dense road network like, for example, Germany, vehicles are probably one of the main threats to all types of wildlife. However, the extent to which this factor affects populations cannot be determined due to a lack of data. This has now been presented by a team of scientists led by the Complutense University of Madrid (UCM), the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) in the journal Perspectives in Ecology and Conservation. The analysis of relevant literature revealed that studies are too limited, involving only a few regions and species. By systematically recording roadkills, the scientists see great potential for assessing the risk of extinction to animal species.
More05.10.2021 | Evolution and Adaptation, sDiv, TOP NEWS
iDiv researcher Alexander Zizka wins GBIF research prize
Based on a media release by GBIF Copenhagen, Leipzig. Today, an international team led by Dr Alexander Zizka of the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University (UL) has earned selection as first place in the Ebbe Nielsen Challenge of the Global Biodiversity Information Facility (GBIF). GBIF is an international network and data infrastructure funded by the world’s governments. Zizka and his team were awarded for developing Bio-Dem, an open-source web-app for exploring temporal and spatial relationships between the availability of biodiversity data and the dimensions of democracy. For its selection, Zizka and his team will receive 12,000 euros.
More22.09.2021 | iDiv Members, Media Release, TOP NEWS
Functioning of terrestrial ecosystems is governed by three main factors
Based on a media release by the Max Planck Institute for Biogeochemistry (MPI BGC) Jena. Ecosystems provide multiple services for humans. However, these services depend on basic ecosystem functions which are shaped by natural conditions like climate and species composition, and human interventions. A large international research team, led by the Max Planck Institute for Biogeochemistry (MPI BGC), and the German Centre for Integrative Biodiversity Research (iDiv), identified three key indicators that together summarise the integrative function of terrestrial ecosystems: The first is the capacity to maximise primary productivity, the second is the efficiency of using water, and the third is the efficiency of using carbon. The monitoring of these key indicators will allow a description of ecosystem function that shapes the ability to adapt, survive and thrive in response to climatic and environmental changes. The study was recently published in the journal Nature.
More17.09.2021 | iDiv, Media Release, Research, TOP NEWS
Species extinction seen also in literature
Leipzig/Frankfurt a.M. Biodiversity has been steadily declining in western literature since the 1830s. This is the finding of a comprehensive, interdisciplinary study led by Leipzig researchers, who examined almost 16,000 works of Western fiction from between 1705 and 1969. The study has been published in the journal People and Nature. The researchers interpret the results as an indication of humanity’s increasing estrangement from nature.
More15.09.2021 | iDiv, Media Release, TOP NEWS
New iDiv research centre building inaugurated
Leipzig. Michael Kretschmer, Bodo Ramelow and Dr Reiner Haseloff – the Ministers President of Saxony, Thuringia and Saxony-Anhalt respectively – together with DFG Secretary General Heide Ahrens, officially opened the new research building of the German Centre for Integrative Biodiversity Research (iDiv) on Wednesday. The event, which took place under strict hygiene measures, was attended by more than 100 guests. They learned about the contribution iDiv is making to solving the global biodiversity crises and intends to make in the future. German Chancellor Dr Angela Merkel sent a message congratulating the iDiv team. The new building at Leipzig’s Alte Messe has been designed as a place for scientists from all over the world to share ideas and conduct integrative research. From 2024 onwards, the three federal states involved intend to take over the financing of the research centre together with other sponsors.
More13.09.2021 | Biodiversity Conservation, Media Release, TOP NEWS
Pioneering method of assessing rewilding progress applied for the first time
Based on a media release by Rewilding Europe Scientists from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and Rewilding Europe have developed a new way of evaluating rewilding progress. Its ground-breaking application across seven of Rewilding Europe’s operational areas has revealed both positive impact and challenges to upscaling. The practical tool can help to inform decision-making and drive rewilding onwards and upwards. The study was recently published in the scientific journal Ecography.
More06.09.2021 | Biodiversity and People, Media Release, TOP NEWS
Large herbivores can reduce fire risks
Leipzig. The use of large herbivores can be an effective means to prevent and mitigate wildfires, especially in places facing land abandonment. They can replace much more costly solutions like firefighting or mechanical vegetation removal. This is the result of a systematic literature review carried out by researchers from the German Centre for Integrative Biodiversity Research (iDiv) and published in the Journal of Applied Ecology. They provide suggestions for fire and agricultural policies in Europe and globally
More31.08.2021 | Evolution and Adaptation, Evolutionary Ecology, iDiv Members, Media Release
To understand future habitat needs for chimpanzees, look to the past
Based on a media release by the Wildlife Conservation Society Leipzig. A new study provides insight into where chimpanzees avoided climate instability during glacial and interglacial periods in Africa over the past 120,000 years. Using bioclimatic variables and other data, the international research team led by the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology (MPI-EVA) and Leipzig University (UL) identified previously unknown swaths of habitat, where chimps rode out the changes seen since the last interglacial period. The findings, published in the American Journal of Primatology, help to increase the understanding of how climate change impacts biodiversity, and how to mitigate against predicted biodiversity loss in the future.
More05.08.2021 | iDiv Members, Media Release, Molecular Interaction Ecology, TOP NEWS
Survival strategy of starving spruces trees: the critical role of reserves
Based on a media release from the Max Planck Institute for Biogeochemistry Young spruce trees build up nutrient reserves even during long periods of famine. This is what scientists from the Max Planck Institute for Biogeochemistry (BGC), the German Centre for Integrative Biodiversity Research (iDiv), the Friedrich Schiller University of Jena (FSU) and other institutions have discovered. They manage to do this even when the energy supply through photosynthesis is paralysed in the long term, as in the case of climate extremes, for example by stopping their growth and even obtaining energy through self-digestion. Until now, it was assumed that this was only possible if enough photosynthesis products were available and growth processes were covered. The study has now been published in the journal PNAS. The knowledge can be used to improve models that predict the fate of forests against the background of climate change.
More13.07.2021 | iDiv Members, Media Release, TOP NEWS
Special honour for biodiversity researcher Josef Settele
New species in the animal and plant kingdoms are often named after public figures. Prof Dr Josef Settele has now received this special honour: A new genus was named after the biodiversity researcher and butterfly expert, who conducts research at the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv): Setteleia. It consists of four moth species that belong to the subfamily of Arctiinae and occur in the Philippines.
More09.07.2021 | Biodiversity Economics, iDiv Members, Media Release, Molecular Interaction Ecology, TOP NEWS
Coastal Ecosystems Worldwide: Billion-dollar Carbon Reservoirs
Based on a media release by the Kiel Institute for the World Economy (IfW Kiel) Kiel/Leipzig. Coastal ecosystems such as seagrass meadows, salt marshes and mangrove forests are valuable to humans in many ways. In particular, they store carbon – and do so with a much higher surface density than forests, for example. They thus make an important contribution to mitigating climate change. Australia’s coastal ecosystems alone, which absorb a particularly large amount of CO2 from the atmosphere, save the rest of the world climate-related costs of around 23 billion US dollar a year. This is according to calculations just published in Nature Climate Change by researchers at the Kiel Institute for the World Economy (IfW Kiel), GEOMAR Helmholtz-Centre for Ocean Research Kiel, the Universities of Kiel and Leipzig (UL) and the German Centre for Integrative Biodiversity Research (iDiv).
More07.07.2021 | Biodiversity and People, iDiv Members, Media Release, TOP NEWS
Alarming state of our streams and rivers
Based on a media release by the Helmholtz Centre for Environmental Research (UFZ) Berlin/Leipzig. The Friends of the Earth Germany (BUND), the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig are launching comprehensive monitoring of German rivers. The scientists want to study the pollution of smaller streams through pesticides and to record the effects on aquatic insect communities. The project aims to investigate the condition of German small water bodies in the long term and to promote biodiversity on this basis. Citizen scientists contribute to this process.
More07.07.2021 | iDiv, Media Release, TOP NEWS
Into the third funding phase with a boost for growth
Bonn. The German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig has succeeded in its application for further funding from the German Research Foundation (DFG). On Tuesday, the Joint Committee of the DFG approved support for iDiv in the third funding phase with around 11.5 million euros annually. This represents an increase of 26 percent over the second funding phase. Even after DFG funding ends, probably in 2024, backing for iDiv as a multipolar university centre for biodiversity research with Central German roots and global purview is to be continued by its host institutions and federal states, along with the support of other funders such as the state of Germany.
More07.07.2021 | iDiv, Media Release, TOP NEWS
The art of biodiversity
Leipzig. There is a new, public space in front of the German Centre for Integrative Biodiversity Research (iDiv) in Leipzig. Located in the entrance area to the Alte Messe old trade fair area, it was created as part of the art in public places Kunst am Bau competition organised by the Staatsbetrieb Sächsisches Immobilien- und Baumanagement (SIB) (Saxon State Land and Construction Management). The artist group nachbars garten and the landscape architecture firm Station C23 were awarded the contract for their joint concept named chorus. The space welcomes visitors with winding paths, flowering lawns and groups of rounded, fusiform figures. It invites you to discover, wonder and linger, and arouses interest in the research centre.
More
22.06.2021 | iDiv Members, Media Release, MLU News, sDiv, TOP NEWS
Vegetation of planet Earth: Researchers publish unique database as Open Access
Based on a media release by the University of Halle-Wittenberg (MLU) Halle/Amiens. It’s a treasure trove of data: the global geodatabase of vegetation plots “sPlotOpen” is now freely accessible. It contains data on vegetation from 114 countries and from all climate zones on Earth. The database was compiled by an international team of researchers led by Martin Luther University Halle-Wittenberg (MLU), the German Centre for Integrative Biodiversity Research (iDiv) and the French National Centre for Scientific Research (CNRS). Researchers around the world finally have a balanced, representative dataset of the Earth’s vegetation at their disposal, as the team reports in the journal Global Ecology & Biogeography.
More18.06.2021 | Biodiversity and People, iDiv, Media Release, Research, TOP NEWS
Dragonflies: Species losses and gains in Germany
Leipzig/Jena. Over the past 35 years, there have been large shifts in the distributions of many dragonfly and damselfly species in Germany. Many species of standing water habitats have declined, probably due to loss of habitat. On the other hand, running-water species, living in rivers and creeks, and warm-adapted species have benefited from improved water quality and warmer temperatures. This was found by a team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Friedrich Schiller University Jena (FSU) and the Helmholtz Centre for Environmental Research (UFZ). The study, published in Diversity and Distributions, highlights the importance of citizen science and natural history societies for long-term data collection and nature conservation efforts for improving biodiversity.
More11.06.2021 | iDiv, Media Release, Species Interaction Ecology, TOP NEWS
Leipzig’s Botanical Garden is humming and buzzing again
Leipzig. From 12 June to 30 September, the Botanical Garden of Leipzig University hosts an exhibition that sheds light on the world of plants and their pollinators. The collaborative project of young researchers and artists from Leipzig shows that there is much more to it than the story of flowers and bees.
More10.06.2021 | iDiv, Media Release, sDiv, TOP NEWS
How climate defines the traits of plant roots
Leipzig/Wyoming/Wageningen. The specific traits of a plant’s roots determine the climatic conditions under which this plant prevails. A new study led by the University of Wyoming (UW) together with the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), and Wageningen University & Research (WUR) published in Nature Ecology and Evolution sheds light on this relationship – and challenges the nature of ecological trade-offs.
More10.06.2021 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
Climate change, biodiversity loss and social justice – these challenges can only be overcome together
Based on a media release by Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research (AWI) and Helmholtz-Centre for Environmental Research (UFZ) Kiel/Leipzig. The fight against global warming and for sustainable development can only succeed if, from now on, humankind considers the issues of climate change, biodiversity loss and social justice together, and takes them into account equally in all political decisions – globally, nationally and regionally – as well as their interactions. According to the German co-authors, this is the most important takeaway from a new workshop report on biodiversity and climate change, the first to be jointly prepared by experts from the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) and the Intergovernmental Panel on Climate Change (IPCC). The report to which scientists from AWI, UFZ, the German Centre for Integrative Biodiversity Research (iDiv) and other institutions contributed, shows why especially phasing out fossil fuels is important for limiting global warming and for conserving nature. In addition, they demonstrate how healthy ecosystems can make long-term contributions to climate.
More07.06.2021 | Evolutionary Ecology, iDiv Members, Media Release, TOP NEWS
Global change: a tight squeeze for African great apes
Based on a media release from the Wildlife Conservation Society Liverpool/Leipzig/Halle. Climate change will drastically reduce the range of African great apes over the next 30 years. This was predicted by an international team of researchers with the participation of the German Centre for Integrative Biodiversity Research (iDiv), the Max Planck Institute for Evolutionary Anthropology and the Martin Luther University Halle-Wittenberg (MLU). In various models, they calculated the effects of climate change, changing land use and human population growth on the future range of gorillas, chimpanzees and bonobos. The existing protected areas are not sufficient to preserve important populations in the long term, the researchers warn in the journal Diversity and Distributions.
More01.06.2021 | iDiv, Media Release, Research, TOP NEWS
Dimensions of invasion success
Based on a media release of the University of Konstanz Invasive alien plants are plant species that grow in an environment outside their native habitat. If they successfully establish self-sustaining populations in these new environments – an event called “naturalization” – they can have considerable negative impacts on local ecosystems, economies, and societies. But not all alien plant species are equally effective in invading new habitats. Therefore, an international team of scientists, including several iDiv members and headed by Konstanz-based biologist Professor Mark van Kleunen, investigated different types of “invasiveness” and possible factors that determine invasion success of alien plants in Europe.
More31.05.2021 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS
Urban life is not to everyone’s taste
Halle/Jena/Leipzig. Rapidly expanding urban habitats are likely to endanger a large number of butterfly species in the long term. This is reported by scientists from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the Friedrich Schiller University Jena (FSU) in the scientific journal Global Change Biology. Generalists that tolerate large temperature fluctuations and feed on many different plants are most likely to benefit from human-modified habitats. In order to preserve biodiversity, urban and spatial planning should take the needs of specialised butterfly species into account, the authors recommend.
More19.05.2021 | iDiv Members, Media Release, TOP NEWS
Palm Oil Plantations Change the Social Behaviour of Macaques
Based on a press release by the University of Leipzig In many parts of Southeast Asia, the rainforest is being replaced by palm oil plantations. Due to the massive clearing of their habitat, some primates move to the plantations in search of food. This often leads to conflicts with farmers, even though the monkeys hardly harm them but, on the contrary, help to keep pests such as rats at low levels. The regular stay on the plantations, however, has a massive influence on the social behaviour of the macaques. This is shown in a new study by scientists from the Max Planck Institute for Evolutionary Anthropology (MPI-EVA), Leipzig University (UL), Universiti Sains Malaysia (USM) and the German Centre for Integrative Biodiversity Research (iDiv). Living in a manmade environment could have a negative effect on the development of the offspring and thus endanger the survival of the macaque populations in the long run. The new findings, recently published in Scientific Reports, may help to develop appropriate measures for protecting the primate species, which is listed as vulnerable by the IUCN, and to promote peaceful coexistence between humans and wild animals.
More18.05.2021 | Biodiversity and People, iDiv Members, Media Release, TOP NEWS
Protection of biological diversity via the Common Agricultural Policy is achievable
Leipzig/Braunschweig/Rostock. In coordination with the EU Commission, a European team of more than 300 scientists has developed recommendations on how the European Union’s future Common Agricultural Policy (CAP) can, within the scope of instruments that have already been adopted, provide a substantial impetus to halt biodiversity loss. The report, written by researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ), the Thünen Institute and the University of Rostock will be presented to the EU Commission in a public online symposium on 19 May.
More13.05.2021 | Biodiversity Conservation, TOP NEWS
Observing biodiversity change from space
Based on a media release by the University of Twente Enschede/Halle. As humans, we’re currently facing two big environmental crises: climate change and biodiversity loss. The first managed to gain a lot of public attention and funding, whereas the latter goes on more slowly in the background. One of the key problems the biodiversity crisis is facing is the few ways to monitor biodiversity. In his recent publication, an international team led by the University of Twente and with the participation of the German Centre for Integrative Biodiversity Research (iDiv) and the Martin-Luther University Halle-Wittenberg, linked existing remote sensing products to so-called essential biodiversity variables (EBVs) to measure biodiversity using satellites. The paper has been published today in the journal Nature Ecology & Evolution.
More11.05.2021 | iDiv, Media Release, Research, TOP NEWS
How smartphones can help detect ecological change
Leipzig/Jena/Ilmenau. Mobile apps like Flora Incognita that allow automated identification of wild plants cannot only identify plant species, but also uncover large-scale ecological patterns. These patterns are surprisingly similar to the ones derived from long-term inventory data of the German flora, even though they have been acquired over much shorter time periods and are influenced by user behaviour. This opens up new perspectives for rapid detection of biodiversity changes. These are the key results of a study led by a team of researchers from Central Germany, which has recently been published in Ecography.
More03.05.2021 | Species Interaction Ecology, Biodiversity Synthesis, iDiv Members, Physiological Diversity, TOP NEWS, UFZ-News
Less precipitation means less plant diversity
Based on a media release by the Helmholtz Centre for Environmental Research (UFZ) Leipzig/Halle. Water is a scarce resource in many of the Earth’s ecosystems. This scarcity is likely to increase in the course of climate change. This, in turn, might lead to a considerable decline in plant diversity. Using experimental data from all over the world, scientists from the Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv), and the Martin Luther University of Halle-Wittenberg (MLU) have demonstrated for the first time that plant biodiversity in drylands is particularly sensitive to changes in precipitation. In an article published in Nature Communications, the team warns that this can also have consequences for the people living in the affected regions.
More27.04.2021 | iDiv, Media Release, TOP NEWS
Limited value of tree plantations for biodiversity conservation
Leipzig/Jena/Halle. Tree plantations are supposed to help compensate the loss of pristine forest habitats. However, their contribution to biodiversity conservation is limited: For example, plantations host a significantly lower number of beetle species as well as individuals than old-growth forests. This was found by a global analysis published in Forest Ecology and Management and led by the German Centre for Integrative Biodiversity Research (iDiv), Friedrich Schiller University Jena (FSU) and Martin Luther University Halle-Wittenberg (MLU).
More12.04.2021 | Experimental Interaction Ecology, iDiv, Media Release, TOP NEWS
Nadia Soudzilovskaia wins prestigous German research prize for international fungi research
Based on a media release of Universiteit Leiden. Environmental scientist Nadia Soudzilovskaia has been awarded the prestigious, international Friedrich Wilhelm Bessel Research Award by the Alexander von Humboldt Foundation. This German award is issued to outstanding foreign mid-career scientists that collaborate with German researchers. ‘The combination of different skills and approaches makes her research highly relevant and innovative!’
More08.04.2021 | Biodiversity Conservation, iDiv, Media Release, Research
Transforming crop and timber production could reduce species extinction risk by 40%
Based on a media release of the International Union for Conservation of Nature (IUCN). Gland (Switzerland). Ensuring sustainability of crop and timber production would mitigate the greatest drivers of terrestrial wildlife decline, responsible for 40% of the overall extinction risk of amphibians, birds and mammals. This is one of the key results of a paper published in Nature Ecology & Evolution. The work was led by the IUCN Species Survival Commission’s Post-2020 Taskforce, hosted by Newcastle University, in collaboration with scientists from 54 institutions in 21 countries around the world, including the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU). The results were generated using a new metric which, for the first time, allows business, governments and civil society to assess their potential contributions to stemming global species loss.
More07.04.2021 | iDiv Members, Media Release, sDiv, Species Interaction Ecology, TOP NEWS
We don’t know how most mammals will respond to climate change
Based on a press release by the British Ecological Society Zurich/Leipzig. A new scientific review has found there are significant gaps in our knowledge of how mammal populations are responding to climate change, particularly in regions most sensitive to climate change. The study was initialised through sDiv, the synthesis centre of the German Centre for Integrative Biodiversity Research (iDiv), involving researchers from the University of Zurich, the Martin Luther University Halle-Wittenberg (MLU), the Helmholtz Centre for Environmental Research (UFZ) and others. The findings are published in the Journal of Animal Ecology.
More31.03.2021 | iDiv, Media Release, Research, TOP NEWS, yDiv
Biodiversity is positively related to mental health
Leipzig/Frankfurt/Kiel. The higher the number of plant and bird species in a region, the healthier the people who live there. This was found by a new study published in Landscape and Urban Planning and led by the German Centre for Integrative Biodiversity Research (iDiv), the Senckenberg Biodiversity and Climate Research Centre (SBiK-F) and the Christian Albrechts University (CAU) in Kiel. The researchers found that, in particular, mental health and higher species diversity are positively related, whereas a similar relationship between plant or bird species and physical health could not be proven.
More23.03.2021 | iDiv Members, Media Release, sDiv, Species Interaction Ecology, TOP NEWS
Short-lived plant species are more climate-sensitive
Plant species with short generation times are more sensitive to climate change than those with long generation times. This is one of the findings of a study by researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ). The international team comprehensively compiled worldwide available data, mostly from Europe and North America, to address the question of how plant populations react to climate change. The study, published in Nature Communications, shows that plant characteristics such as generation time can predict how sensitive species are to changing climates. This has important implications for predicting which plant species need the most conservation attention regarding climate change.
More17.03.2021 | iDiv Members, Media Release, TOP NEWS
New method for genome assembly in barley provides excellent results
Based on a press release by IPK Leibniz Institute Gatersleben. Genomes differ between the individuals of one species and we can learn a lot about diversity in our crops by comparing genomes of different varieties. However, researchers that want to study many genomes need a fast and reliable method for sequence assembly. An international research team led by the IPK Leibniz Institute and the German Centre for Integrative Biodiversity Research (iDiv) has now investigated a new DNA sequencing method. The results, which have now been published in the journal The Plant Cell, are very promising. The scientists now hope to be able to use the method for assembling other barley genomes in the future.
More05.03.2021 | iDiv Members, Media Release, Sustainability and Complexity in Ape Habitat, TOP NEWS
Chimpanzees without borders
Based on a press release by the Max Planck Institute for Evolutionary Anthropology (MPI-EVA) Chimpanzees are divided into four subspecies separated by geographic barriers like rivers. Previous studies attempting to understand chimpanzee population histories have drawn inconsistent pictures: some have shown clear separations between chimpanzee subspecies while others suggest a genetic gradient across the species as in humans. To resolve this dichotomy, a team of international researchers led by the Pan African Programme: The Cultured Chimpanzee (PanAf), the Max Planck Institute for Evolutionary Anthropology and the German Centre for Integrative Biodiversity Research (iDiv) collected over 5000 faecal samples from 55 sites in 18 countries across the chimpanzee range over 8 years – the most complete sampling of the species to date. The analysis was published in Communications Biology.
More05.03.2021 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS
Species are our livelihoods
In future, assessments of ecosystems in terms of their contributions to people should take greater account of the importance of species diversity. This is what scientists from the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) are calling for in the scientific journal Ecosystem Services. Animal and plant species deliver a large share of natural services for human well-being but have been in continuous decline for decades. Large-scale assessments, however, only address some of these services, such as water filtration, carbon storage and erosion control. Ecosystem services that are directly linked to species, on the other hand, hardly play a role at all. The researchers point out that this wastes many opportunities for effective nature and species conservation.
More24.02.2021 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
Belowground biodiversity in motion
Based on a press release by the Max Planck Institute for Mathematics in the Sciences Global change is expected to increase the diversity of bacteria at the local level, while their composition will become more uniform at the global level. This is the finding of a team of researchers led by the German Center for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU), the University of Leipzig (UL) and the Max Planck Institute for Mathematics in the Sciences (MiS). The researchers have – for the first time – holistically assessed how climate and land-use changes affect bacterial communities in soil. The study, published in Global Ecology and Biogeography, can help make reliable predictions about changes in the global distribution of these belowground communities.
More23.02.2021 | iDiv Members, sDiv, Theory in Biodiversity Science, TOP NEWS
Complex river ecosystems protect freshwater fish from species loss
Report by Dr Stefano Larsen, Fondazione Edmund Mach, Italy Trento/Leipzig/Seattle. The conservation and restoration of highly branched river courses can help to curb the extinction of fish species. This is the result of an international working group led by Edmund Mach Foundation and with participation of the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich Schiller University Jena (FSU), supported by iDiv’s synthesis centre sDiv. The researchers found that temporal synchrony in the abundance of freshwater fish populations can be predicted directly from the branching complexity and connectivity of river networks. Synchronous fluctuations in the size of different populations of a species at different locations in a river significantly increase the probability of extinction. As species are lost from freshwaters faster than in any other ecosystems, knowledge of fundamental patterns in their spatial and temporal dynamics can inform conservation strategies both now and in the future. The study was recently published in Ecology Letters.
More22.02.2021 | iDiv, Media Release, Species Interaction Ecology, TOP NEWS
Plant responses to climate are lagged
Leipzig. Plant responses to climate drivers such as temperature and precipitation may become visible only years after the actual climate event. This is a key result of new research led by the German Centre of Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ) published in Global Change Biology. The results indicate that climate drivers may have different effects on the survivorship, growth and reproduction of plant species than suggested by earlier studies.
More22.02.2021 | iDiv Members, sDiv, TOP NEWS
Can species adapt to a hotter world?
Based on a press release by Lancaster University, UK Species evolve faster to adapt to the climate getting colder than to it getting hotter – and the rise in heat they can adapt to has limits. These are the main results of a study by an international team led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg, the University Rey Juan Carlos and University of Alcalá in Spain, involving researchers from Lancaster University and the Helmholtz Centre for Environmental Research (UFZ). The new study, published in Nature Communications, explores how evolution has prepared some species to withstand these temperatures and how fast different species evolve to changing temperatures.
More09.02.2021 | Biodiversity and People, iDiv Members, TOP NEWS
Why biological diversity is important for our health
Report by first author Dr Melissa Marselle, former post-doctoral researcher at UFZ/iDiv, and senior author Prof Aletta Bonn, head of ecosystem services lab at UFZ/iDiv/FSU. Leipzig. Biodiversity has a major impact on human well-being – in four different ways: by reducing harm, restoring capacities, building capacities and causing harm. This is the result of a team of 26 international experts from various disciplines, led by the Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv), as well as Friedrich Schiller University Jena (FSU). The study was recently published in the journal Environment International.
More26.01.2021 | iDiv Members, Media Release, TOP NEWS
Autumn and spring are closely linked
Based on a media release by Friedrich Schiller Universität Jena The fact that plants are beginning to flower earlier and earlier as a result of climate change was reported some time ago by scientists in Jena, among others. But how do the climatic changes actually affect the other end of the growing season? To find answers to this question, biologists Dr Solveig Franziska Bucher of the Friedrich Schiller University Jena and Prof Christine Roemermann of the Friedrich Schiller University Jena and the German Centre for Integrative Biodiversity Research (iDiv) have intensively studied the so-called leaf senescence, i.e. the ageing process of plants, which can be observed, for example, through autumnal colouring or the shedding of leaves. They discovered that leaf senescence begins earlier at lower temperatures than at higher temperatures. The start of this process can differ from species to species, but the colder the environment, the faster it occurs. The study was published in the Journal of Ecology.
More26.01.2021 | Biodiversity Conservation, iDiv Members, Media Release, TOP NEWS
Avoid repeating old mistakes
Leipzig/Halle. In the future, global goals for biodiversity must apply to all member states of the UN Convention on Biological Diversity (CBD) also at national level. This is one of four recommendations for improving the global strategy for biodiversity made by a team of researchers led by the Nanjing Institute for Environmental Research in China, the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and NatureServe. The global biodiversity strategy is currently being renegotiated under the auspices of the CBD. In addition, funding, knowledge base and methods for measuring the success of nature conservation should be improved. In the journal Nature Ecology & Evolution, the researchers analyse why the global goals for biodiversity have been largely missed so far and present concrete policy options.
More25.01.2021 | Biodiversity and People, iDiv Members, Media Release, TOP NEWS
Street trees close to the home may reduce the risk of depression
Leipzig. Daily contact with trees in the street may significantly reduce the risk of depression and the need for antidepressants. This is the result of a study by researchers at the Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), and the Friedrich Schiller University Jena (FSU), recently published in the journal Scientific Reports. Street tree planting in residential areas of cities can serve as a nature-based solution to reduce the risk of depression with added benefits of also addressing climate change and biodiversity loss. This should be taken into account by urban planners, health professionals, and conservationists.
More14.01.2021 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
Measuring the belowground world
Leipzig/Halle/Fort Collins. A quarter of all known species live in the soil. Life above ground depends on the soil and its countless inhabitants. Yet, global strategies to protect biodiversity have so far paid little attention to this habitat. In the journal Science, an international team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU), Leipzig University (UL) and Colorado State University calls for greater consideration of soils in the renegotiation of international biodiversity strategies. Their relevance must be recognised far beyond agriculture. In order to make the status and performance of soils more visible, the researchers explain their plan for systematic recording based on common global standards.
More04.01.2021 | iDiv, TOP NEWS
“The Global Biodiversity Agenda and the European Biodiversity Strategy: Opportunities and Challenges for Central Germany”
With the Ministers of the Environment of Saxony-Anhalt, Thuringia and Saxony, Prof. Dr. Claudia Dalbert, Anja Siegesmund and Wolfram Günther, as well as the President of the Federal Agency for Nature Conservation Prof. Dr. Beate Jessel and several iDiv scientists.
More14.12.2020 | iDiv Members, Media Release, sDiv, TOP NEWS
Plant diversity in Germany on the decline
Bonn/Halle/Jena/Leipzig. Germany’s plant diversity is on the decline: over the last 60 years, decreases have been observed across Germany in over 70 percent of the more than 2000 species examined. The species dropped by an average of 15 percent. These are the findings of the most comprehensive analysis of plant data from Germany ever conducted, recently published in Global Change Biology. 29 million pieces of data on the distribution of vascular plants were taken into account in the analyses carried out within the framework of the “sMon – Biodiversity Trends in Germany” project of the German Centre for Integrative Biodiversity Research (iDiv). The study involved researchers from iDiv, the universities of Jena, Halle and Rostock, the Helmholtz Centre for Environmental Research (UFZ) and the Federal Agency for Nature Conservation (BfN) with the close involvement of the regional nature conservation agencies from all 16 federal states.
More10.12.2020 | Experimental Interaction Ecology, Media Release, TOP NEWS
Leipzig biodiversity researcher receives 2021 Leibniz Prize
Leipzig. Today, the Joint Committee of the DFG (Deutsche Forschungsgemeinschaft) awarded the 2021 Gottfried Wilhelm Leibniz Prize to four female and six male researchers. One of them is Nico Eisenhauer, research group head at the German Centre for Integrative Biodiversity Research (iDiv) and Professor at Leipzig University since 2014 . The awards are endowed with 2.5 million euros each.
More03.12.2020 | Biodiversity and People, iDiv Members, Media Release, TOP NEWS, yDiv
Does biodiversity evoke happiness?
Based on a media release by Senckenberg. Frankfurt, Leipzig, Kiel. A high biological diversity in our immediate vicinity is as important for life satisfaction as our income. These are the findings that scientists from Senckenberg, the German Centre for Integrative Biodiversity Research (iDiv), and the University of Kiel recently published in the journal Ecological Economics. For the first time, the researchers were able to show that all across Europe, the individual enjoyment of life is correlated to the number of bird species in one’s surroundings. An additional ten percent of bird species in the vicinity therefore increases the life satisfaction of Europeans at least as much as a comparable increase in income. Nature conservation thus constitutes an investment in human well-being, according to the researchers.
More26.11.2020 | iDiv Members, Media Release, TOP NEWS
Leipzig researchers compile world’s largest inventory of known plant species
Leipzig. Researchers at Leipzig University (UL) and the German Centre for Integrative Biodiversity Research (iDiv) have compiled the world’s most comprehensive list of known plant species. It contains 1,315,562 names of vascular plants, thus extending the number of recognised plant species and subspecies by some 70,000 – equivalent to about 20%. The researchers have also succeeded in clarifying 181,000 hitherto unclear species names. The data set has now been published in Scientific Data. This marks the culmination of more than ten years of intensive research work and could help to make Leipzig a leading international centre of plant biodiversity research.
More26.11.2020 | Biodiversity Conservation, Media Release, TOP NEWS
EuropaBON: MLU and iDiv lead new pan-European project for joint monitoring
Based on a media release by the Martin Luther University Halle-Wittenberg (MLU) A new research project aims at developing a transnational system for monitoring biodiversity and ecosystems in Europe. It is being coordinated by the Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig. The results of the project are to inform the political decisions of the European Commission and other decision-makers. The EU is providing three million euros in funding for the project. A virtual kick-off meeting of the project partners will take place from 1 to 3 December.
More19.11.2020 | TOP NEWS
Highly Cited Researchers 2020
Clarivate Analytics lists 12 iDiv members in its 2020 selection of “Highly Cited Researchers”. According to Clarivate Analytics, these scientists have demonstrated significant influence through publication of multiple papers, highly cited by their peers, during the last decade.
More06.11.2020 | Experimental Interaction Ecology, iDiv, Theory in Biodiversity Science, TOP NEWS
More plant diversity, less pesticides
Leipzig/Jena/Minnesota. Increasing plant diversity enhances the natural control of insect herbivory in grasslands. Species-rich plant communities support natural predators and simultaneously provide less valuable food for herbivores. This was found by a team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv), who conducted two analogous experiments in Germany and the USA. Their results were published in Science Advances and show that increasing plant biodiversity could help reduce pesticide inputs in agricultural systems by enhancing natural biological control.
More29.10.2020 | Evolutionary Ecology, Media Release, TOP NEWS
The seductive scent of sweet fruits
The German Centre for Integrative Biodiversity Research (iDiv) adds a new research field to its portfolio. From November 2020, the biologist Dr Omer Nevo will head the new Junior Research Group ‘Evolutionary Ecology’ at iDiv and Friedrich Schiller University Jena (FSU). The 35-year-old scientist is particularly interested in chemical communication between plants and animals – in detail, how plants attract seed-dispersing animals by their fruit scent and how both sides have adapted to each other. This knowledge about the signalling effect of odours should also help to find new approaches in nature conservation. Nevo and his team will be funded for six years with about 1.3 million euros from the Emmy Noether Programme of the Deutsche Forschungsgemeinschaft (DFG).
More28.10.2020 | iDiv Members, Media Release, sDiv, TOP NEWS
Climate change drives plants to extinction in the Black Forest in Germany, study finds
Based on a press release by the Martin Luther University Halle-Wittenberg (MLU) Climate change is leaving its mark on the bog complexes of the German Black Forest. Due to rising temperatures and longer dry periods, two plant species have already gone extinct over the last 40 years. The populations of many others have decreased by one third. In the next couple of decades ten more species could become extinct, researchers from Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) write in Diversity and Distributions.
More27.10.2020 | Biodiversity and People, Media Release, sDiv, Sustainability and Complexity in Ape Habitat, TOP NEWS
Biodiversity monitoring programmes need a culture of collaboration
Biodiversity loss is continuing relentlessly worldwide. In order to counteract this more effectively, monitoring programmes are needed which precisely map the circumstances of animal and plant species and the extent to which they are under threat. But too often, these are still inadequate – the range of species examined is not extensive enough, and there is too little coordination. A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv) describes, in an article for the scientific journal One Earth how different stakeholders can combine their data and expertise to improve monitoring and thus counteract further species loss.
More26.10.2020 | Biodiversity and People, Media Release, TOP NEWS
Land management in forest and grasslands: how much can we intensify?
Based on a media release by the University of Bern Leipzig/Bern. High land-use intensity reduces the beneficial effects of biodiversity on ecosystem services. This is the main result of a study conducted by an international team led by researchers from the Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv) and the University of Bern. The study published in PNAS assessed, for the first time, the effects of land management on the links between three ecosystem attributes simultaniously: biodiversity, ecosystem functions and ecosystem services. It identified thresholds of management intensity, where these relationships change dramatically, which species groups were most important in driving services, and the ecosystem services that are at risk when management is intensified.
More26.10.2020 | Experimental Interaction Ecology, iDiv, iDiv Members, Media Release, Research, TOP NEWS
Shifts in flowering phases of plants due to reduced insect density
Based on a media release by the Friedrich Schiller University Jena Leipzig/Jena. It still sounds unlikely today, but declines in insect numbers could well make it a frequent occurrence in the future: fields full of flowers, but not a bee in sight. A research group of the University of Jena and the German Centre for Integrative Biodiversity Research (iDiv) has discovered that insects have a decisive influence on the biodiversity and flowering phases of plants. If there is a lack of insects where the plants are growing, their flowering behaviour changes. This can result in the lifecycles of the insects and the flowering periods of the plants no longer coinciding. If the insects seek nectar at the wrong time, some plants will no longer be pollinated.
More23.10.2020 | Biodiversity Conservation, Media Release, TOP NEWS
We need a safety net for biodiversity
Based on a media release by the National University of Córdoba and the Leibniz-Institute of Freshwater Ecology and Inland Fisheries (IGB) Córdoba, Leipzig, Halle. A “safety net” made up of multiple interlinked and ambitious goals is needed to tackle nature’s alarming decline. No single goal can capture the broad range of characteristics that need to be sustained, concludes a large international team with the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the Leibniz-Institute of Freshwater Ecology and Inland Fisheries (IGB). The study, published in the journal Science, outlines the scientific basis for redesigning the new set of biodiversity goals.
More19.10.2020 | iDiv Members, Media Release, TOP NEWS
Academies demand rapid action to preserve biodiversity in the agricultural landscape
More07.10.2020 | Experimental Interaction Ecology, iDiv Members, Media Release, TOP NEWS
Long-term consequences of global change difficult to predict
Based on a media release by Leipzig University In a longitudinal study, an international research team led by Leipzig University (UL) and the German Centre for Integrative Biodiversity Research (iDiv) has investigated the consequences of changes in plant biodiversity for the functioning of ecosystems. The scientists found that the relationships between plant traits and ecosystem functions change from year to year. This makes predicting the long-term consequences of biodiversity change extremely difficult, they write in Nature Ecology & Evolution.
More06.10.2020 | Media Release, TOP NEWS
Efficient pollen identification
Based on a media release by the Helmholtz Centre for Environmental Research (UFZ) From pollen forecasting, honey analysis and climate-related changes in plant-pollinator interactions, analysing pollen plays an important role in many areas of research. Microscopy is still the gold standard, but it is very time consuming and requires considerable expertise. In cooperation with Technische Universität (TU) Ilmenau, scientists from the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv) have now developed a method that allows them to efficiently automate the process of pollen analysis. Their study has been published in the specialist journal New Phytologist.
More29.09.2020 | Biodiversity Synthesis, iDiv, Media Release, Research, TOP NEWS
‘Cool’ sampling sites more likely to show false trends in biodiversity change
Leipzig/Halle. Data collected by citizen science initiatives, museums and national parks is an important basis for research on biodiversity change. However, scientists from the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) found that sampling sites are oftentimes not representative, which may lead to false conclusions about how biodiversity changes. Their research, published in Conservation Biology, calls for more objective site selection and better training for citizen scientists to prevent a site-selection bias.
More28.09.2020 | Evolution and Adaptation, iDiv, Media Release, sDiv, Species Interaction Ecology, TOP NEWS
Artificial intelligence can help protect orchids and other species
Leipzig/Halle. Orchids may be decorative, but many orchid species are also threatened by land conversion and illegal harvesting. However, only a fraction of those species is included in the IUCN Red List of Threatened Species, because assessments require a lot of time, resources and expertise. A new approach, an automated assessment developed under the lead of biodiversity researchers from Central Germany, now shows that almost 30% of all orchid species are possibly threatened. The findings have been published in Conservation Biology and suggest that the new approach could speed up conservation assessments of all species on Earth.
More15.09.2020 | Media Release, Research, TOP NEWS
Europe’s primary forests: What to protect? What to restore?
Halle/Berlin. An expansion of the protected areas by only about 1% would sufficiently protect most remaining primary forests in Europe. This is one of the main results of a study conducted by an international team led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU) and Humboldt-Universität zu Berlin (HU). The study, published in Diversity and Distribution, is the first assessment of the conservation status of Europe’s primary forests. It identifies protection gaps and areas with restoration needs to reach conservation targets. In addition, it provides valuable information how to implement the new EU Biodiversity Strategy.
More14.09.2020 | Media Release, Sustainability and Complexity in Ape Habitat, TOP NEWS
Chimpanzees show greater behavioural and cultural diversity in more variable environments
Based on a media release by the Max Planck Institute for Evolutionary Anthropology (MPI EVA) Leipzig. Chimpanzee behavioural and cultural diversity has been well documented across equatorial Africa, however, the ecological-evolutionary mechanisms are not yet understood. An international team led by the Max Planck Institute for Evolutionary Anthropology and the German Centre for Integrative Biodiversity Research (iDiv) has investigated the influence of environmental variability on the behavioural repertoires of 144 social groups. The study, published in Nature Communications, found that chimpanzees living further away from historical forest refugia, under more seasonal conditions, and found in savannah woodland rather than closed forested habitats, were more likely to exhibit a larger set of behaviours.
More09.09.2020 | Macroecology and Society, Media Release, TOP NEWS
Bending the curve of biodiversity loss
Based on a press release by the International Institute for Applied Systems Analysis (IIASA) Laxenburg/Leipzig. Ambitious, integrated action combining conservation and restoration efforts with a transformation of the food system. This is the recipe for turning the tide of biodiversity loss by 2050 or earlier, a new study led by the International Institute for Applied Systems Analysis (IIASA) with participation of researchers from the German Centre for Integrative Biodiversity Research (iDiv) suggests. Using multiple models and scenarios, the team investigated whether and how it might be feasible to improve the status of biodiversity while at the same time complying with other sustainability targets related to land use. The study was published in Nature and forms part of the latest WWF Living Planet Report.
More03.09.2020 | iDiv, iDiv Members, Media Release, TOP NEWS
ERC Starting Grant for iDiv junior scientists Dr Martin Mascher
Based on a media release by the Leibniz Institute for Plant Genetics and Crop Plant Research (IPK) Gatersleben: Great success for iDiv Junior Research Group head Dr Martin Mascher. The 34 years old scientist affiliated to the Leibniz Institute for Plant Genetics and Crop Plant Research (IPK) receives a Starting Grant from the European Research Council (ERC). His current research project TRANSFER will thus receive 1.5 million euros in funding over the next five years.
More01.09.2020 | Biodiversity and People, Media Release, TOP NEWS, UFZ-News
Awareness raising alone is not enough
Based on a media release by the Helmholtz Centre for Environmental Research (UFZ) It is a well-known problem: too rarely do nature conservation initiatives, recommendations or strategies announced by politicians lead to people really changing their everyday behaviour. A German-Israeli research team led by the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv) has investigated the reasons for this. According to the team, the measures proposed by politicians do not sufficiently exploit the range of possible behavioural interventions and too rarely specify the actual target groups, they write in the journal Conservation Biology.
More31.08.2020 | Biodiversity and People, iDiv Members, TOP NEWS, yDiv
Contributions of wildlife to human well-being in the scientific literature gaining in importance
Report by Joel Methorst (iDiv, Senckenberg, Goethe University Frankfurt), yDiv doctoral researcher Leipzig, Frankfurt. Non-material contributions of wildlife (WCP) to human well-being are gaining attention in the scientific literature. Whereas until the end of the 20th century only negative evaluations such as health hazards were mentioned, the focus on positive influences on physical and mental health as well as on culture aspects are now becoming increasingly popular in research. These are the main findings of a new study conducted by scientists at the German Centre for Integrative Biodiversity Research (iDiv), Senckenberg Biodiversity and Climate Research Centre (SBiK-F) and Goethe University Frankfurt. The study was published in Environmental Research Letters.
More28.08.2020 | iDiv, Media Release, TOP NEWS
Leipzig’s floodplain suffering from chronic disease
Leipzig. Leipzig’s floodplain forest is one of the largest of its kind in Central Europe. But the floodplain is currently under huge pressure – as a result not only of drought and climate change but also of human intervention. These stressors as well as revitalization measures for the large urban forest were discussed during a visit to Leipzig’s Canopy Crane. Among the participants were Professor Beate Jessel from the German Federal Agency for Nature Conservation (BfN), Wolfram Günther from the Environmental Ministry of Saxony, Rüdiger Dittmar from the City of Leipzig and Professor Christian Wirth from the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University (UL).
More26.08.2020 | Biodiversity Conservation, Media Release, Sustainability and Complexity in Ape Habitat, TOP NEWS
Effectiveness of primate conservation measures mostly unproved
Leipzig/Halle/Cambridge. Less than one percent of scientific literature on primates evaluate the effectiveness of interventions for the conservation of primates. That is the result of a new study compiled by a team of experts in 21 countries, led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU), Max Planck Institute for Evolutionary Anthropology (MPI-EVA) and the University of Cambridge. Despite great protection efforts, this is one of the main reasons for the dramatic decline of the populations of primate species. The study published in the journal BioScience proposes several actions to improve the evidence base for the conservation of primates.
More24.08.2020 | Experimental Interaction Ecology, iDiv, iDiv Members, Media Release, Research, TOP NEWS
Ecologists put biodiversity experiments to the test
Based on a media release of the Senckenberg Biodiversity and Climate Research Centre Leipzig/Frankfurt/Bern. Much of our knowledge of how biodiversity benefits ecosystems comes from experimental sites. These sites contain combinations of species that are not found in the real world, which has led some ecologists to question the findings from biodiversity experiments. But the positive effects of biodiversity for the functioning of ecosystems are more than an artefact of experimental design. This is the result of a new study led by an international team of researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), the University of Bern and the Senckenberg Biodiversity and Climate Research Centre. For their study, they removed ‘unrealistic’ communities from the analysis of data from two large-scale experiments. The results that have now been published in Nature Ecology & Evolution show that previous findings are, indeed, reliable.
More17.08.2020 | iDiv, iDiv Members, Media Release, MLU News, Research, TOP NEWS
Does city life make bumblebees larger?
Based on a media release of the Martin Luther University Halle-Wittenberg. Leipzig/Halle. Does urbanisation drive bumblebee evolution? A new study by Martin Luther University Halle-Wittenberg (MLU) and the German Centre for Integrative Biodiversity Research (iDiv) provides an initial indication of this. According to the study, bumblebees are larger in cities and, therefore, more productive than their rural counterparts. In Evolutionary Applications, the research team reports that differences in body size maybe caused by the increasingly fragmented habitats in cities.
More17.08.2020 | iDiv, iDiv Members, MLU News, Physiological Diversity, Research, TOP NEWS, UFZ-News
Are tipping points suitable concepts for developing environmental policies?
Based on an media release of Carl von Ossietzky University Oldenburg Oldenburg/Leipzig/Halle. Many policies tackling the consequences of global environmental change rely on the concept of tipping points: If an impact, such as biodiversity loss, becomes too large, an ecosystem might flip into a different often less desirable state. However, a new study published in Nature Ecology and Evolution and led by an international team of scientists from the University of Oldenburg, the German Centre for Integrative Biodiversity Research (iDiv) and other research institutions finds little evidence for thresholds. When focussing on tipping points, scientists and policy makers may thus risk overlooking the negative impact of gradual changes on ecosystems – with potentially disastrous consequences.
More10.08.2020 | iDiv, Media Release, MLU News, Research, sDiv, Species Interaction Ecology, TOP NEWS, UFZ-News
Land-use change disrupts wild plant pollination
Leipzig/Halle. Human changes to the environment have been linked to widespread pollinator declines. New research published in Nature Communications shows that intensive land use will further decrease pollination and reproductive success of wild plants, especially of those plants that are highly specialised in their pollination. An international team of scientists led by researches from the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ) performed a global data analysis that provided conclusive evidence of the links of human land use and pollination of plants.
More04.08.2020 | Experimental Interaction Ecology, Media Release, TOP NEWS
Chemicals inhibit decomposition processes – by damaging biodiversity
Leipzig, Namur. Declines in the diversity and abundance of decomposers explain reductions in plant decay rates under the influence of chemical stressors, but not added nutrients. These are the new insights of a study published in the open access journal eLife. The global meta-analysis conducted by researchers at the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL) and the University of Namur in Belgium highlights the main anthropogenic effects on the biodiversity and functioning of ecosystems, and thus helps predicting the fate of different ecosystems around the world.
More03.08.2020 | Experimental Interaction Ecology, iDiv, Media Release, MLU News, Research, TOP NEWS
Identifying the blind spots of soil biodiversity
Leipzig/Halle. Soils harbour a substantial part of the world’s biodiversity, yet data on the patterns and processes taking place below ground does not represent all relevant ecosystems and taxa. For example, tropical and subtropical regions largely remain a blind spot when it comes to soil biodiversity. This is one of the results of a new study published in Nature Communications and led by scientists from the German Centre for Integrative Biodiversity Research, the Martin Luther University Halle-Wittenberg and Leipzig University.
More29.07.2020 | Biodiversity Synthesis, iDiv, Media Release, MLU News, Research, Species Interaction Ecology, TOP NEWS, UFZ-News
Smaller habitats worse than expected for biodiversity
Leipzig/Halle. Biodiversity’s ongoing global decline has prompted policies to protect and restore habitats to minimize animal and plant extinctions. However, biodiversity forecasts used to inform these policies are usually based on assumptions of a simple theoretical model describing how the number of species changes with the amount of habitat. A new study published in the journal Nature shows that the application of this theoretical model underestimates how many species go locally extinct when habitats are lost. Scientists from the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ) used data from 123 studies from across the world to set the path for the next generation of biodiversity forecasts in the face of habitat loss and restoration.
More28.07.2020 | Experimental Interaction Ecology, iDiv Members, TOP NEWS, UFZ-News
Shrinking dwarves
Based on a press release by the Helmholtz Centre for Environmental Research (UFZ) Nowadays, life in the soil must contend with several problems at once. The biomass of small animals that decompose plants in the soil and thus maintain its fertility is declining both as a result of climate change and over-intensive cultivation. To their surprise, however, scientists from the Helmholtz Centre for Environmental Research (UFZ), the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University have discovered that this effect occurs in two different ways: while the changing climate reduces the body size of the organisms, cultivation reduces their frequency. Even by farming organically, it is not possible to counteract all negative consequences of climate change, the researchers warn in the trade journal eLife.
More17.07.2020 | iDiv, Media Release, Research, Sustainability and Complexity in Ape Habitat, TOP NEWS
Clear strategies needed to reduce bushmeat hunting
Leipzig. Extensive wildlife trade not only threatens species worldwide but can also lead to the transmission of zoonotic diseases. It encompasses hundreds of species with significant differences in their conservation status and associated disease risk. However, current strategies to mitigate the wildlife trade often neglect these differences. An international research team led by the German Centre for Integrative Biodiversity Research (iDiv) and the Max Planck Institute for Evolutionary Anthropology (MPI-EVA) shed new light on the motivations why people hunt, trade or consume different species. The research has been published in the journal People and Nature and shows that more differentiated solutions are needed to prevent uncontrolled disease emergence and species decline.
More17.07.2020 | iDiv, Media Release, Species Interaction Ecology, TOP NEWS
Exhibition in Leipzig puts a spotlight on pollination networks
Leipzig. A new exhibition at the Botanical Garden of Leipzig University gives new and unexpected insights into the world of plants and their pollinators. The collaborative project of young researchers and artists from Leipzig shows that there is much more to it than the story of flowers and bees.
More14.07.2020 | iDiv Members, Media Release, TOP NEWS
How many invasive species can our ecosystems tolerate?
Based on a press release by the University of Vienna Even an increase in invasive alien species by 20 to 30 percent will lead to a dramatic global loss of biodiversity in the future. This is the conclusion of a study conducted by an international team of researchers led by the University of Vienna with contributions from the German Centre for Integrative Biodiversity Research (iDiv) and the Helmholtz Centre for Environmental Research (UFZ). The current increase is mainly driven by the global transport of goods, climate change and the extent of economic growth. The study was published in the journal Global Change Biology.
More03.07.2020 | iDiv, iDiv Members, TOP NEWS
Nature data as a basis for decisionmaking in politics and society
Based on a press release of the Friedrich Schiller University of Jena The German Research Foundation (DFG) has approved the application of a nationwide consortium of biodiversity researchers to fund a National Research Data Infrastructure (NFDI) of biodiversity data. The aim is to provide high-quality data, tools and infrastructures that follow the FAIR guiding principles (findability, accessibility, interoperability and reuse) in a sustainable manner. NFDI4BioDiversity is partly coordinated by researchers from the Friedrich Schiller University (FSU), the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ), and many other iDiv partner organisations participate.
More02.07.2020 | iDiv Members, sDiv, TOP NEWS
Root Economics – Between Do-It-Yourself Strategies and Fungal Outsourcing
Based on a press release by Freie Universität Berlin The formation of a plant’s root growth often depends on symbiosis with fungi. This is the main finding of an international group of researchers with members from Freie Universität Berlin (FU), the German Center for Integrative Biodiversity Research (iDiv), and Wageningen University, among others, has been studying the complex belowground economy of roots. In the journal Science Advances the biologists describe the different growth strategies found in root systems and how they “collaborate” with fungi. Their findings are based on the analysis of approximately 1,800 plant species from around the world and the specific traits of their roots. The new insights will help scientists better understand “root economics” and the adaptations and exchanges that take place belowground as well as how environmental changes affect these relationships.
More29.06.2020 | iDiv Members, TOP NEWS, UFZ-News
Scientists warn of increasing threats posed by invasive alien species
Based on a media release by Helmholtz Centre for Environmental Research (UFZ) In a new study, scientists from around the world with participation of researchers from the Centre for Environmental Research (UFZ) and German Centre for Integrative Biodiversity Research (iDiv) warn that the threats posed by invasive alien species are increasing. They say that urgent action is required in order to prevent, detect, and control invaders at both local and global levels.
More18.06.2020 | Biodiversity Synthesis, iDiv, Media Release, MLU News, Research, TOP NEWS
Forest loss escalates biodiversity change
Based on a media release of the University of St Andrews Forest loss does not always lead to a decline in biodiversity but can amplify how it changes. This is one of the results of a recent study from an international team of researchers, including the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU). Focussing on biodiversity data spanning 150 years and over 6,000 locations, the study, published in the journal Science, reveals that as tree cover is lost across the world’s forests, plants and animals are responding to the transformation of their natural habitats. However, these responses may be delayed by decades.
More15.06.2020 | Biodiversity Synthesis, TOP NEWS
Simulations: personal responsibility alone makes COVID-19 pandemic in Germany manageable – albeit at a high price
Leipzig, Hamburg, Kiel. How high would the infection and death rates be if Germany had relied solely on voluntary social distancing at the beginning of the pandemic? A new study by the German Centre for Integrative Biodiversity Research (iDiv) and the universities of Leipzig, Hamburg and Kiel shows that even purely voluntary social distancing – in order to protect ourselves and others – could stabilise the number of new infections (R0=1). However, this would mean 100 times more infections and 15 times as many deaths compared to an optimal policy of reducing social contacts. The study was recently published on a preprint server.
More04.06.2020 | Biodiversity and People, iDiv, Media Release, TOP NEWS
Citizen Science Dialogue Forum
This event will be held in German language only. Please refer to the German press release.
More25.05.2020 | Biodiversity and People, iDiv Members, Media Release, TOP NEWS, UFZ-News
Turning the spotlight on insects
Based on a media release by Helmholtz Centre for Environmental Research (UFZ) Paris, Halle, Leipzig. The European Union’s (EU) network of Natura 2000 conservation areas is designed to protect endangered animal and plant species and their habitats. But it also benefits a large number of non-target species. However, these beneficiaries are unevenly distributed across the major animal groups, reports an international team of researchers, including biologists from the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv), in the journal Conservation Biology. According to the research team, almost half of the bird species not named in the target group benefit from Natura 2000 protection, but only one forth of the butterflies.
More20.05.2020 | Biodiversity Conservation, Media Release, TOP NEWS
International Day of Biodiversity 2020: the pandemic calls for nature conservation
Leipzig, Halle. 2020 was declared a political “super year” for nature conservation. In October, the Conference of the Parties (COP) to the UN Convention on Biological Diversity (CBD) should have set the course for international biodiversity policies for the next 10 to 30 years. The corona pandemic has pressed the pause button also in this so-called post-2020 process. COP-15 has been postponed until further notice. However, the pandemic also highlights the important role natural ecosystems play for human health – as a source of pathogens, but also as part of the solution. This is what Prof Dr Henrique Miguel Pereira stresses at this year’s International Day of Biodiversity on 22 May themed “Our solutions are in nature”. Pereira is head of the research group Biodiversity and Nature Conservation at the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg and co-chair of the Group on Earth Observations Biodiversity Observation Network (GEO BON). He is involved in several forums with policy makers, including the Intergovernmental Platform on Biodiversity and Ecosystem Services and the CBD.
More12.05.2020 | iDiv, iDiv Members, Media Release, Research, TOP NEWS
Medicinal plants thrive in biodiversity hotspots
Based on a press release of Leipzig University With their rich repertoire of anti-infective substances, medicinal plants have always been key in the human fight to survive pathogens and parasites. This is why the search for herbal drugs with novel structures and effects is still one of the great challenges of natural product research today. Scientists from Leipzig University (UL), the Leibniz Institute of Plant Biochemistry (IPB) and the German Centre for Integrative Biodiversity Research (iDiv) have now shown a way to considerably simplify this search for bioactive natural compounds using data analyses on the phylogenetic relationships, spatial distribution and secondary metabolites of plants. Their new approach makes it possible to predict which groups of plants and which geographical areas are likely to have a particularly high density of species with medicinal effects. This could pave the way for a more targeted search for new medicinal plants in the future.
More11.05.2020 | Experimental Interaction Ecology, Media Release, sDiv, TOP NEWS
More soil-borne plant pathogens in a warmer world
Based on a media release by Pablo de Olavide University Sevilla Sevilla / Leipzig / Halle. Global warming will increase the proportion of plant pathogens in soils across the globe. Many of these plant pathogens also affect important food and medicinal plants, which is likely to have a long-term impact on the world population’s food security. This is the result of an experimental study published in the journal Nature Climate Change with significant contribution by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL) and Martin Luther University Halle-Wittenberg (MLU).
More04.05.2020 | Biodiversity Synthesis, iDiv, Media Release, MLU News, Research, TOP NEWS
Climate change impact greater on marine systems
Based on a media release by the University of St Andrews Temperature changes lead to clear biodiversity responses in marine systems, where warming coincided with increases in the number of species in most locations. These increases in the number of species are likely caused by the influx of climate migrants from species-rich warmer regions. These are the results of a new research, led by scientists from the Universities of Helsinki, St Andrews and Radboud, in collaboration with researchers from the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU). published in Nature Ecology and Evolution and focussed on linking biodiversity change to temperature change, and on comparing responses between ocean and land. The research was published in Nature Ecology and Evolution and focussed on linking biodiversity change to temperature change, and on comparing responses between ocean and land.
More24.04.2020 | Biodiversity Synthesis, Media Release, sDiv, TOP NEWS
Insects: Largest study to date confirms declines on land, but finds recoveries in freshwater
Leipzig. A worldwide compilation of long-term insect abundance studies shows that the number of land-dwelling insects is in decline. On average, there is a global decrease of 0.92% per year, which translates to approximately 24% over 30 years. At the same time, the number of insects living in freshwater, such as midges and mayflies, has increased on average by 1.08% each year. This is possibly due to effective water protection policies. Despite these overall averages, local trends are highly variable, and areas that have been less impacted by humans appear to have weaker trends. These are the results from the largest study of insect change to date, including 1676 sites across the world, now published in the journal Science. The study was led by researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL) and Martin Luther University Halle-Wittenberg (MLU). It fills key knowledge gaps in the context of the much-discussed issue of “insect declines“.
More13.04.2020 | Biodiversity Conservation, iDiv, Media Release, MLU News, Research, sDiv, TOP NEWS
Plant diversity in European forests is declining
Leipzig / Halle. In Europe’s temperate forests, less common plant species are being replaced by more widespread species. An international team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) has found that this development could be related to an increased nitrogen deposition. Their results have been published in the journal Nature Ecology & Evolution.
More09.04.2020 | Computational Forest Ecology, iDiv, Media Release, Research, TOP NEWS
A glimpse into the future of tropical forests
Leipzig / Panama City. Tropical forests are a hotspot of biodiversity. Against the backdrop of climate change, their protection plays a special role and it is important to predict how such diverse forests may change over decades and even centuries. This is exactly what researchers at the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL) and other international research institutions have achieved. Their results have been published in the scientific journal Science.
More18.03.2020 | Biodiversity Conservation, Media Release, TOP NEWS
EU Biodiversity Strategy: Call for nature restoration
More13.03.2020 | iDiv, Media Release, Research, sDiv, TOP NEWS
Plant life on the edge
Edinburgh/Leipzig. All of plant life can be summarised by what shapes plants take and how they grow. An international team of scientists funded by the German Centre for Integrative Biodiversity Research (iDiv) and the Natural Environment Research Council of the UK found that tundra plants use a wide variety of adaptations to deal with the very short summers and long harsh winters found at Arctic latitudes and at the top of mountains. The findings, published in Nature Communication, indicate that tundra plants are more diverse in how they cope with cold climates than previously thought. In a warming world, these tundra plants will benefit from having a wide range of ways to adapt to the changing climate.
More12.03.2020 | Biodiversity and People, sDiv, TOP NEWS
Getting hit from all sides: Anthropogenic threats gang up on biodiversity
Report by Diana Bowler, postdoctoral researcher at the Ecosystem Services group at iDiv, FSU resp. UFZ and first author of a new publications in ‘people and nature’ Leipzig. An important step in assessing which species are most at risk of extinction is determining which places are most affected by human activities. Environmental changes caused by harmful human activities are often grouped as climate change, habitat loss, exploitation (e.g., hunting and fishing), pollution, and spread of non-native species. In this study, we examined maps of the intensities of these human-caused environmental changes across the world. We analysed the spatial relationships among the intensities of these environmental changes to identify which were most associated with each other.
More09.03.2020 | iDiv, iDiv Members, TOP NEWS
TRY database for plant traits has released more than a billion records
More03.03.2020 | Biodiversity and People, iDiv, Media Release, TOP NEWS
More taxpayers’ money for the environment and public benefit
Leipzig. Scientists from across Europe call for swift and effective action from the EU with regard to its Common Agricultural Policy (CAP). In a position paper written by 21 authors and coordinated by scientists from the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ) and the University of Rostock, the EU Commission’s current reform proposals were identified as inadequate. Billions of euros of taxpayers’ money are about to be poured down the drain – making it unlikely to reach the set climate and nature conservation targets or the envisaged social objectives. The researchers are proposing ten measures for a sustainable and fair agricultural policy. Over 3,600 signatories support the call for action.
More26.02.2020 | Evolution and Adaptation, iDiv, Media Release, Research, TOP NEWS
Colour vision in primates closely linked to palm fruite colours
Leipzig/Amsterdam. The evolution of colour vision might be closely linked to the availability of food. Researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL), and the University of Amsterdam (UvA) found that colour vision in African primate species, which is similar to that of humans, is related to the spatial distribution of palm fruit colours. The results of their study have been published in Proceedings of the Royal Society B. They shed new light on the evolution of primates.
More14.02.2020 | iDiv, Media Release, Research, sDiv, TOP NEWS
Climate change will lead to abrupt shifts in dryland ecosystems
Based on a media release of the University of Alicante Alicante. Increases in aridity can alter the capacity of dryland ecosystems to sustain life, also limiting the provision of essential ecosystem services to more than 2 billion people living in those areas, such as soil fertility and biomass production. This was shown by a study recently published in Science. The study was led by Fernando T. Maestre from the University of Alicante, who is also a sabbatical at sDiv, the synthesis centre of the German Centre for Integrative Biodiversity Research (iDiv).
More31.01.2020 | iDiv, Media Release, Research, sDiv, TOP NEWS
Global science team on red alert as Arctic lands grow greener
Based on a media release of the University of Edinburgh New research techniques including drone and satellite technology are being adopted by scientists tackling the most visible impact of climate change – the so-called greening of Arctic regions. A team of 40 scientists from 36 institutions, supported by the German Centre for Integrative Biodiversity Research (iDiv), revealed that the causes of this greening process are more complex – and variable – than was previously thought. Their findings have now been published in Nature Climate Change.
More29.01.2020 | iDiv Members, Media Release, TOP NEWS, yDiv
Pollination is better in cities than in the countryside
Flowering plants are better pollinated in urban than in rural areas. This has now been demonstrated experimentally by a team of scientists led by the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the Helmholtz Centre for Environmental Research (UFZ). Although the scientists found a greater diversity of flying insects in the countryside, more bees in cities resulted in more pollinated flowers of test plants. By far the most industrious pollinators were bumble bees, most likely benefitting from the abundant habitats available in the city. To promote pollination, the researchers recommend to take into greater account the needs of bees when landscape planning – both in cities and in the countryside. Their results have been published in the journal Nature Communications.
More23.01.2020 | Biodiversity and People, iDiv, Media Release, TOP NEWS
Public lecture: Butterflies and Politics
More22.01.2020 | Media Release, sDiv, TOP NEWS
Even after death, animals are important in ecosystems
Animal carcasses play an important role in biodiversity and the functioning of ecosystems, also over prolonged periods. Scientists from the German Centre for Integrative Biodiversity Research (iDiv) and the University of Groningen have published these findings in the journal PLOS ONE. The carcasses not only provide food for many carrion-eating animal species, their nutrients also contribute to the significantly increased growth of surrounding plants. This, in turn, attracts many herbivorous insects and their predators. The researchers recommend relaxing regulations governing the disposal of animal carcasses when applied to nature conservation areas.
More17.01.2020 | Biodiversity Conservation, iDiv, Media Release, MLU News, TOP NEWS
Bringing back nature to the EU in the post-2020 Biodiversity Strategy
Based on a media release of Rewilding Europe The rewilding of European ecosystems can help to tackle both the current climate and biodiversity emergencies. In a policy brief published today, experts from six organisations, including the German Centre of Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU), call on the European Commission to prioritise nature recovery in the EU Biodiversity Strategy post-2020.
More13.01.2020 | iDiv, Media Release, sDiv, TOP NEWS
iDiv receives award by the United Nations Decade on Biodiversity
More17.12.2019 | iDiv, TOP NEWS
Forstwirtschaftsplan: Conservation of species in Leipzig riparian forest needs management
More13.12.2019 | iDiv Members, TOP NEWS, UFZ-News
The Evidence is Clear: Transformative Change Needed Now to
Address Nature Crisis and Protect Human Quality of Life
Press release by the Secretariat of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) on 12 December 2019
More09.12.2019 | Biodiversity Conservation, iDiv, Media Release, Research, sDiv, TOP NEWS
Urban growth causes more biodiversity loss outside of cities
Leipzig/Halle/Arlington. In a rapidly urbanising world, the conversion of natural habitats into urban areas leads to a significant loss of biodiversity in cities. However, these direct effects of urban growth seem to be much smaller than the indirect effects outside of cities, such as the urban release of greenhouse gases causing climate change globally or the increasing demand for food and resources in cities leading to land-use change in rural areas. Both climate and land-use change are key drivers of global biodiversity loss. An international team of researchers including researchers from The Nature Conservancy (TNC), the German Centre for Integrative Biodiversity Research (iDiv), Martin Luther University Halle-Wittenberg (MLU) and other institutions assessed the direct and indirect effects on a global scale. The results have been published in the journal Nature Sustainability.
More22.11.2019 | iDiv, Media Release, Physiological Diversity, Research, sDiv, TOP NEWS
Plant diversity struggles in wake of agricultural abandonment
Based on a media release of the University of Minnesota. Minnesota/Leipzig. Decades after farmland was abandoned, plant diversity and productivity struggle to recover. This has been shown by a new research, published in the journal Nature Ecology & Evolution. Researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ) and the University of Minnesota examined plant diversity and plant productivity on fields that had been ploughed and abandoned for agricultural use. Although local grassland plant diversity increased over time, plant productivity did not significantly recover.
More08.11.2019 | Biodiversity and People, iDiv Members, Media Release
Plant species with medium abundance have declined the most.
More24.10.2019 | Experimental Interaction Ecology, Media Release, sDiv, TOP NEWS
Higher local earthworm diversity in Europe than in the tropics
Leipzig. In any single location, there are typically more earthworms and more earthworm species found in temperate regions than in the tropics. Global climate change could lead to significant shifts in earthworm communities worldwide, threatening the many functions they provide. These are the two main results of a new study published in Science. The research was led by scientists from the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University. They brought together 140 researchers from across the globe to compile the largest earthworm dataset worldwide, encompassing 6928 sites in 57 countries.
More21.10.2019 | iDiv, iDiv Members, Media Release, Research, TOP NEWS
Macaques can make palm oil more sustainable and efficient by hunting plantation rats
Leipzig/Gelugor. In Malaysia, wild pig-tailed macaques do not have the best reputation and are even considered a crop pest. Contrary to this, they actually feed on rats, the major oil palm pest, and can provide an important ecosystem service as biological pest control agent. This was found by a team of researchers from Universiti Sains Malaysia (USM), the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University (UL) and the Max Planck Institute for Evolutionary Anthropology (MPI EVA). Their research investigates the costs and benefits of macaques foraging in oil palm plantations and was published in Current Biology.
More18.10.2019 | Biodiversity Synthesis, iDiv, Media Release, Research, sDiv, TOP NEWS
The composition of species is changing in ecosystems across the globe
Leipzig/Halle/St Andrews. Biodiversity is changing rapidly in many places all over the world. However, while the identities of species in local assemblages are undergoing significant changes, the number of species is on average remaining relatively constant. Thus, changes in local assemblages do not always reflect the species losses occurring at the global scale. These findings are based on observations by a team of scientists led by the German Centre for Integrative Biodiversity Research (iDiv), the Martin Luther University Halle-Wittenberg (MLU) and the University of St Andrews. Their findings, which have been published in Science, reveal greater changes in species composition in marine systems than on land, with most extreme changes in tropical marine biomes.
More08.10.2019 | Biodiversity Conservation, iDiv, Media Release, Research, TOP NEWS
Global South most affected by climate change and land use effects
Based on a media release of the Natural Capital Project Leipzig/Stanford. By 2050, up to 5 billion people could be at higher risk of water pollution, coastal storms or under-pollinated crops with a majority living in developing countries. This is one of the most alarming results of a study recently published in Science by an international research team with contribution of the German Centre of Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU). The team produced a high-resolution global map showing the distribution of nature’s ability to provide services to humankind. These services are increasingly threatened by human-driven degradation of ecosystems and biodiversity. The map may improve policy and decision-making around investments in nature locally as well as globally. With the help of sustainable development, the threats to vital ecosystem services may be reduced.
More30.09.2019 | Experimental Interaction Ecology, iDiv, Media Release, Research, TOP NEWS
Building bridges in Central Germany
Joint media release of iDiv, the University of Leipzig and the Friedrich Schiller University Jena Jena/Leipzig. The Deutsche Forschungsgemeinschaft (DFG) funds a new research unit at the Jena Experiment with a total of 5 million euros for five years. The research will be lead by the German Centre for Integrative Biodiversity Research (iDiv), the University of Leipzig and the Friedrich Schiller University in Jena and will investigate the mechanisms underlying the relation between biodiversity and ecosystem functions. For this purpose, the new research unit will also make use of other research platforms.
More26.09.2019 | Biodiversity Economics, Media Release, TOP NEWS
German fishermen’s scepticism towards EU impedes compliance with its regulations
Leipzig/Hamburg/Kiel: Negative perception of a regulatory authority like the EU diminishes the honesty of those regulated, for example, that of fishermen. This is the conclusion drawn by researchers from the German Centre for Integrative Biodiversity Research (iDiv), the Leipzig University, the University of Hamburg and the Kiel Institute for the World Economy from a game of chance experiment with EU-sceptic commercial fishermen and Brexit voters. Among other things, the findings, published in the European Economic Review, help to assess the effectiveness of unmonitored EU fisheries regulations. The experiment also revealed that the fishermen were more honest than students.
More18.09.2019 | iDiv, Media Release, sDiv, TOP NEWS
Climate and biodiversity go hand in hand – climate change destroys our natural resources
Leipzig. Global warming is changing nature and its diversity. It puts the resources at risk, that our diet and fundamental aspects of human life rely on. Increasing scarcity of clean water and fertile soil may lead to global conflicts. Many employees from the German Centre for Integrative Biodiversity Research (iDiv) will participate in the Fridays for Future demonstration which takes place on 20 September in Leipzig to get this on top of the political agenda. iDiv reseachers Nicole van Dam and Roel van Klink plan to give statements at Leipzig’s Augustusplatz.
More16.09.2019 | Media Release, Species Interaction Ecology, TOP NEWS
Study: We need more realistic experiments on the impact of climate change on ecosystems
Based on a Media release by Martin Luther University Halle-Wittenberg (MLU) When it comes to the impact of climate change on ecosystems, we still have large knowledge gaps. Most experiments are unrealistic because they do not correspond to projected climate scenarios for a specific region. As a result, we lack reliable data on what ecosystems might look like in the future, as a team of biodiversity researchers from Central Germany show in the journal Global Change Biology. The team reviewed all experimental studies on the topic. The researchers are now calling for the introduction of common protocols for future experiments.
More16.09.2019 | Media Release, Species Interaction Ecology, TOP NEWS
Study: We need more realistic experiments on the impact of climate change on ecosystems
Based on a Media release by Martin Luther University Halle-Wittenberg (MLU) When it comes to the impact of climate change on ecosystems, we still have large knowledge gaps. Most experiments are unrealistic because they do not correspond to projected climate scenarios for a specific region. As a result, we lack reliable data on what ecosystems might look like in the future, as a team of biodiversity researchers from Central Germany show in the journal Global Change Biology. The team reviewed all experimental studies on the topic. The researchers are now calling for the introduction of common protocols for future experiments.
More05.09.2019 | iDiv, Media Release, TOP NEWS
Diversity increases ecosystem stability
Based on a media release by the University of Freiburg Leipzig/Freiburg: Forests with a large variety of species are more productive and stable under stress than monocultures: scientists from the University of Freiburg and the German Centre for Integrative Biodiversity Research (iDiv) have confirmed this with data from the world’s oldest field trial on the diversity of tropical tree species. The team around doctoral researcher Florian Schnabel has published its results in the journal Global Change Biology.
More02.08.2019 | Biodiversity and People, Media Release, sDiv, TOP NEWS
EU agriculture not viable for the future
Leipzig, Brussels. The current reform proposals of the EU Commission on the Common Agricultural Policy (CAP) are unlikely to improve environmental protection, say researchers led by the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ) and the University of Göttingen in the journal Science. While the EU has committed to greater sustainability, this is not reflected in the CAP reform proposal. The authors show how the ongoing reform process could still accommodate conclusive scientific findings and public demand to address environmental challenges including climate change.
More29.07.2019 | Biodiversity Synthesis, Media Release, TOP NEWS
Introduced species dilute the effects of evolution on diversity
International research team investigates mechanisms of forest biodiversity in Hawaiian archipelago Media release by the University of Göttingen Göttingen/Halle/Leipzig/Hawaii: Understanding how biodiversity is shaped by multiple forces is crucial to protect rare species and unique ecosystems. Now an international research team led by the University of Göttingen, the German Centre for Integrative Biodiversity Research (iDiv), the Helmholtz Centre for Environmental Research (UFZ) and the Martin Luther University Halle-Wittenberg (MLU) together with the University of Hawai’i at Mānoa has found that biodiversity is higher on older islands than on younger ones. Furthermore, they found that introduced species are diluting the effects of island age on patterns of local biodiversity. The findings were published in PNAS (Proceedings of the National Academy of Sciences).
More09.04.2019 | iDiv Members, Media Release, TOP NEWS
Plant diversity increases insect diversity
Halle, Leipzig, Göttingen. The more plant species live in grasslands and forests, the more insect species find a habitat there. However, the presence of more plant species does not only increase the number of insect species, but also the number of insect individuals. Simultaneously, animal diversity is not only determined by plant diversity, but also by the physical structure of the plant communities. These are the results of an international collaboration led by the German Centre for Integrative Biodiversity Research (iDiv), published in the journal Nature Communications. These results have consequences for the insect-friendly management of grasslands and forests.
More02.04.2019 | Biodiversity Synthesis, iDiv, Media Release, TOP NEWS
Loss of habitat causes double damage to species richness
Halle (GER), Lunz (A). Loss and fragmentation of habitat are among the main reasons why biodiversity is decreasing in many places worldwide. Now, a research team with participation of the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) has established that the destruction of habitat causes double damage to biodiversity; if habitat patches disappear, not only do the species living there become extinct, but species richness in neighbouring patches also declines. The reason for this additional species loss is the large physical distances between the remaining habitat patches, the researchers write in the journal Ecology Letters.
More27.03.2019 | Biodiversity and People, Media Release, TOP NEWS
Europe’s farmlands are losing their insect-eating birds
Frankfurt am Main/Leipzig. The number of birds that primarily feed on insects has decreased significantly during the past 25 years in the European Union. This is mainly due to the decline of these so-called insectivores in agricultural landscapes, researchers from the German Senckenberg Biodiversity and Climate Research Centre and the German Centre for Integrative Biodiversity Research (iDiv) report currently in the scientific journal Conservation Biology. It is the first Europe-wide study that examines how population trends of birds are related to their diet. Insectivores make up roughly half of all European bird species.
More13.03.2019 | Biodiversity Conservation, Experimental Interaction Ecology, iDiv Members, Media Release, sDiv, TOP NEWS
Mismatches between above- and belowground biodiversity
Leipzig. After conducting comprehensive studies, an international team of researchers led by Leipzig University and the iDiv research centre has gained important new insights into biodiversity above and below the earth’s surface: they discovered that, on about 30 per cent of our planet’s terrestrial surface, there is great diversity of flora and fauna in the soil compared to considerably fewer species above the ground, or vice versa, biodiversity is much greater above than below ground. These differences in biodiversity were not detectable in the remaining 70 per cent of the earth’s terrestrial surface. Here there were hot and cold spots that displayed either high or low overall levels of biodiversity. The researchers have published the results of their study in the journal Conservation Biology.
More04.03.2019 | Biodiversity Conservation, Media Release, TOP NEWS
Biodiversity crisis: Technological advances in agriculture are not a sufficient response
Leipzig, Halle. Rapid population and economic growth are destroying biological diversity – especially in the tropics. This was reported by a research team led by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) in Nature Ecology & Evolution. A constantly growing demand for agricultural products requires ever new cultivated areas. Even though technological advances are making agriculture ever more efficient, the growing number of people makes up for these successes. The study shows: an effective nature conservation policy needs concepts against population growth and for sustainable consumption.
More20.02.2019 | Biodiversity Synthesis, Media Release, TOP NEWS
Complete world map of tree diversity
Leipzig, Halle. The biodiversity of our planet is one of our most precious resources. However, for most places in the world, we only have a tiny picture of what this diversity actually is. Researchers at the German Centre for Integrative Biodiversity Research (iDiv) and Martin Luther University Halle-Wittenberg (MLU) have now succeeded in constructing, from scattered data, a world map of biodiversity showing numbers of tree species. With the new map, the researchers were able to infer what drives the global distribution of tree species richness. Climate plays a central role; however, the number of species that can be found in a specific region also depends on the spatial scale of the observation, the researchers report in the journal Nature Ecology and Evolution. The new approach could help to improve global conservation.
More21.01.2019 | Experimental Interaction Ecology, Media Release, sDiv, TOP NEWS
Courage to aim for less cleanliness?
Leipzig, Raleigh. Do the same laws of biodiversity which apply in nature also apply to our own bodies and homes? If so, current hygiene measures to combat aggressive germs could be, to some extent, counterproductive. So writes an interdisciplinary team of researchers from the German Centre for Integrative Biodiversity Research (iDiv), Leipzig University and North Carolina State University in the journal Nature Ecology & Evolution. They propose that examination of the role diversity of microorganisms plays in the ecosystems of our bodies and homes should be intensified. The findings could challenge existing strategies for fighting infectious diseases and resistant germs.
More05.11.2018 | Media Release, Sustainability and Complexity in Ape Habitat, TOP NEWS
Despite government claims, orangutan populations have not increased
Leipzig/Brunei. Orangutan populations are still declining rapidly, despite claims by the Indonesian Government that things are looking better for the red apes. In the journal Current Biology, a team of scientists including Maria Voigt of the German Centre for Integrative Biodiversity Research (iDiv) and the Max Planck Institute for Evolutionary Anthropology criticise the use of inappropriate methods for assessing management impacts on wildlife trends. The researchers call for scientifically sound measures to be employed in order to ensure that wildlife monitoring provides reliable numbers.
More01.10.2018 | iDiv, Media Release, Research, TOP NEWS
Species-rich forests store twice as much carbon as monocultures
Halle (Saale). Species-rich subtropical forests can take up, on average, twice as much carbon as monocultures. This has been reported by an international research team in the professional journal Science. The study was carried out as part of a unique field experiment conducted under the direction of Martin Luther University Halle-Wittenberg (MLU), the German Centre for Integrative Biodiversity Research (iDiv) and the Chinese Academy of Sciences. The experiment comprises forests grown specifically for this purpose in China; for the study, data from experimental plots with a total of over 150,000 trees were analysed. The researchers believe that the results speak in favour of using many different tree species during reforestation. Thus, both species conservation and climate protection can be promoted.
More27.09.2018 | Biodiversity Economics, iDiv, Media Release, TOP NEWS
New Professor for Biodiversity Economics: Martin Quaas completes iDiv research centre
Leipzig. The German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig and the Faculty of Economics and Management Science at Leipzig University will receive a new joint professorship: Martin Quaas will join the faculty as Professor of Biodiversity Economics on 1 October. Quaas researches interactions between economics and biodiversity; for example, the effect of quotas on fishermen’s incomes and stocks of marine fish species. Quaas completes a group of nine professors specifically appointed to develop the iDiv research centre, which is funded by the DFG and was founded in 2012.
More26.09.2018 | Computational Forest Ecology, Media Release, sDiv, TOP NEWS
New plants on the block: Taller species are taking over in a warming Arctic
Frankfurt/ Leipzig, Germany, 24.09.2018. Until now, the Arctic tundra has been the domain of low-growing grasses and dwarf shrubs. Defying the harsh conditions, these plants huddle close to the ground and often grow only a few centimetres high. But new, taller plant species have been slowly taking over this chilly neighbourhood, report an international group of nearly 130 biologists led by scientists from the German Centre for Integrative Biodiversity Research (iDiv) and the German Senckenberg Biodiversity and Climate Research Centre today in Nature. This has led to an overall increase in the height of tundra plant communities over the past three decades.
More06.08.2018 | iDiv, Media Release, TOP NEWS
“Frankfurter Erklärung” on the protection of biodiversity
More01.08.2018 | iDiv Members, Media Release, TOP NEWS
Animals and fungi foster forest multifunctionality
Leipzig, Halle. A new study shows that, in addition to the diversity of tree species, the variety of animal and fungus species also has a decisive influence on the performance of forests. Forest performance comprises many facets besides timber production, such as carbon storage and climate regulation. The study is based on ten years of research in species-rich subtropical forests. A team of researchers led by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin-Luther-University Halle-Wittenberg has published the results in the new issue of Nature Communications. They illustrate that biodiversity must be viewed as a whole in order to maintain the performance of forests.
More24.07.2018 | Biodiversity Conservation, Media Release, TOP NEWS
Plenty of habitat for bears in Europe
Leipzig/Halle. Great opportunity for European brown bears: a new study spearheaded by the German Centre for Integrative Biodiversity Research (iDiv) and the Martin Luther University Halle-Wittenberg (MLU) shows that there are still many areas in Europe where bears are extinct but with suitable habitat for hosting the species. An effective management of the species, including a reduction of direct pressures by humans (like hunting), has the potential to help these animals come back in many of these areas, according to the head of the study. It is now important to plan the recovery of the species while taking measures to prevent conflicts.
More04.07.2018 | Experimental Interaction Ecology, Media Release, TOP NEWS
Slug shuttle-service for mites
Leipzig. On the menu for slugs are not only mosses, lichens and garden vegetables, but also miniscule oribatid mites, which they unavoidably take in with their food. Astonishingly, most of these tiny arachnids survive the voyage through the slug’s digestive system without harm and are excreted, alive, elsewhere in the ecosystem. Scientists led by Dr Manfred Türke from the German Centre for Integrative Biodiversity Research (iDiv) and Leipzig University have, for the first time, discovered this dispersal strategy, used predominantly by plants and known in the scientific community as endozoochory (dispersal by ingestion), also being used by mites. The researchers have published their findings in the journal Oecologia.
More04.06.2018 | Evolution and Adaptation, Media Release, TOP NEWS
iDiv research centre continues to grow – Dutch evolutionary biologist sets up new junior research group
Leipzig. The German Centre for Integrative Biodiversity Research (iDiv) continues to grow. As of June, the Dutch biologist Dr Renske Emilie Onstein will head up a new “Evolution and Adaptation” junior research group with offices and laboratories based in Leipzig’s Bio City. Using flowering plants and fruit-eating animals, the botanist and her team will investigate how species originate and which factors influence this process. This knowledge is important to predict how biological diversity might adapt to global change.
More15.05.2018 | Biodiversity Conservation, Media Release, TOP NEWS
New “Silk Road” brings challenges and opportunities for biodiversity conservation
London. In an article published this week by the renowned journal Nature Sustainability, an international team of scientists argues that environmental protection should be a priority for the “Belt and Road” initiative. This Chinese project would then represent not only an investment to foster international trade but also an opportunity for sustainable development leadership. Among the team, who calls for rigorous strategic environmental and social assessments, are researchers from the Research Center in Biodiversity and Genetic Resources (CIBIO-InBIO) in Portugal, the Martin Luther University Halle-Wittenberg and the German Centre for Integrative Biodiversity Research (iDiv).
More26.04.2018 | Media Release, Sustainability and Complexity in Ape Habitat, TOP NEWS
Gorillas and chimpanzees: more than expected, but in danger
Based on a media release by the Wildlife Conservation Society (WCS): New York/Leipzig. A study estimates that more than 360,000 gorillas and nearly 130,000 chimpanzees still inhabit the forests of Africa approximately – one third and one tenth more than previously thought. However, approximately 80 percent of these great apes live outside protected areas, and gorillas are declining at an annual rate of 2.7 percent. Efforts to stop poaching, illegal logging, and habitat degradation and destruction are key to saving great apes. Conservationists from several organizations and government agencies gathered and analysed data on western lowland gorilla and central chimpanzee populations in the largest ever survey of these great apes that live exclusively in Western equatorial Africa. The field work for the study collectively took 167 person-years, with the researchers walking a distance longer than the north-south axis of Africa. The study that has been published in the journal Science Advances was led by the Wildlife Conservation Society (WCS). Among the authors is Hjalmar Kühl from the iDiv research centre and the Max Planck Institute for Evolutionary Anthropology.
More09.03.2018 | Biodiversity Conservation, Media Release, TOP NEWS
The lynx is back in the Thuringian Forest
Leipzig/Oberschönau. With the support of a local nature conservationist, scientists from the iDiv research center and the Martin Luther University Halle-Wittenberg have found evidence for the presence of the lynx in the Thuringian Forest. North of the village of Oberschönau an adult individual was photographed by a camera trap. The photos proof unequivocally that the shy carnivore is back in the Thuringian Forest – 200 years after its dissapearance.
More05.03.2018 | iDiv Members, Media Release, TOP NEWS
Common media release by iDiv, UFZ and Senckenberg
Scientists agree: The time to act is now
As part of a hearing in Saxony’s State Parliament on the causes of insect decline on March 2, 2018, scientists from iDiv, Senckenberg and UFZ provided their expertise, next to representatives of agriculture and other organizations. The scientists unanimously stressed the need to act now, especially since both the facts of insect decline and its main causes are undisputed.
More21.12.2017 | Experimental Interaction Ecology, Media Release, TOP NEWS
Climate change: Self-enhancing effect cannot be explained by soil animals
Leipzig. When the soil warms up, it releases more carbon dioxide (CO2) – an effect that further fuels climate change. Until now, it had been assumed that the reason for this was mainly due to the presence of small soil animals and microorganisms that would eat and breathe more in warmer temperatures. However, a new study in Nature Climate Change has shown that this is not the case. Quite the contrary: If warmth is accompanied by drought, the soil animals eat even less. In order to improve the predictive power of climate models, it is now crucial to understand the biological processes in the soil better, say the scientists.
More17.11.2017 | iDiv Members, Media Release, Research, TOP NEWS
The importance of biodiversity in forests could increase due to climate change
Leipzig. Forests fulfil numerous important functions, and do so particularly well if they are rich in different species of trees. This is the result of a new study. In addition, forest managers do not have to decide on the provision of solely one service – such as wood production or nature conservation – as a second study demonstrates: several services provided by forest ecosystems can be improved at the same time. Both studies were led by scientists from Leipzig University and the German Centre for Integrative Biodiversity Research (iDiv), and published in the prestigious journal Ecology Letters.
More17.07.2017 | Media Release, Theory in Biodiversity Science, TOP NEWS
Why Tyrannosaurus was a slow runner
A beetle is slower than a mouse, which is slower than a rabbit, which is slower than a cheetah… which is slower than an elephant? No! No other animal on land is faster than a cheetah – the elephant is indeed larger, but slower. For small to medium-sized animals, larger also means faster, but for really large animals, when it comes to speed, everything goes downhill again. For the first time, it is now possible to describe how this parabola-like relationship between body size and speed comes about. A research team under the direction of the German Centre for Integrative Biodiversity Research (iDiv) and the Friedrich-Schiller-University Jena have managed to do so thanks to a new mathematical model, and also published their findings in the prestigious journal ‘Nature Ecology and Evolution’.
More