Biodiversity encompasses multiple levels, scaling from ecosystems down to genetic variation. Ecosystems host diverse communities of interacting species, each contributing to the complex web of life. Zooming in, populations of individual species display genetic diversity, critical for adaptation and resilience. This diversity stems from natural allelic variants — distinct versions of genes within a species’ genome. These alleles encode different proteoforms, or structurally and functionally distinct protein variants, which impact an organism’s traits and ecological interactions, thereby driving evolutionary processes and shaping responses to environmental changes. Understanding biodiversity at the scale of proteoforms is the focus of the recently DFG-funded Collaborative Research Centre 1664 (CRC 1664) – Plant Proteoform Diversity (SNP2Prot).
The SNP2Prot initiative aims to understand how genome-encoded sequence variation translates into structural, mechanistic, and functional proteoform diversity. Traditionally, functional genome analysis and protein biochemistry have been separate fields. The CRC 1664 fosters interdisciplinary collaboration between plant, protein scientists, and computational biologists to uncover how genetic variations influence phenotypic traits. In the long term, this knowledge will guide the design of proteoforms to impact plant traits, aiding plant breeding and molecular pharming.
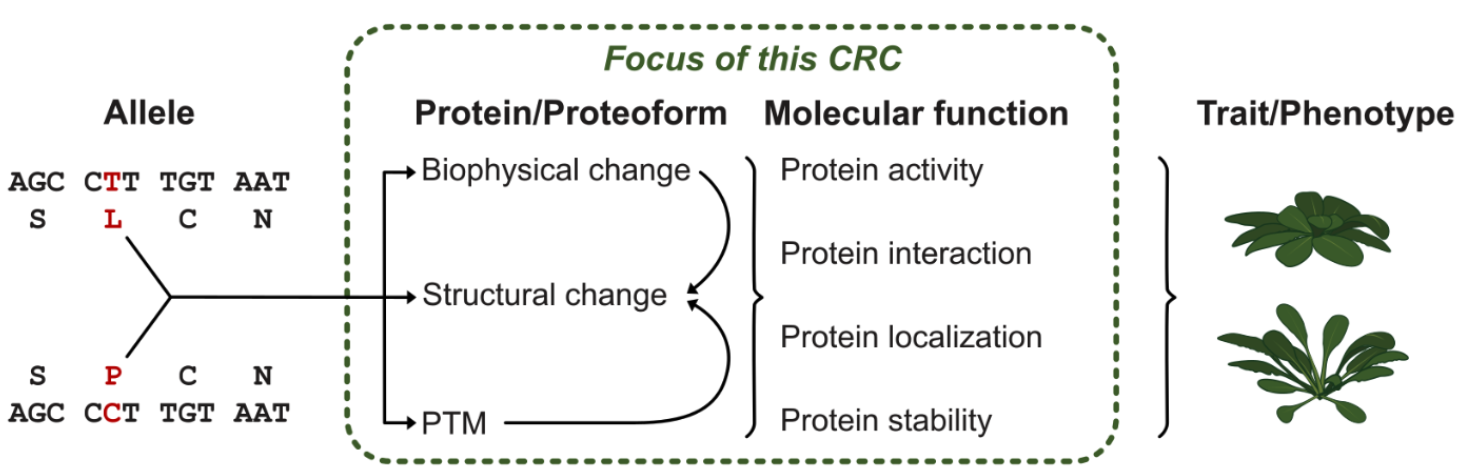

SNP2Prot offers a strong environment for innovative biodiversity research at the level of proteoform variation within plant populations. MLU and iDiv members Ivo Grosse, Sascha Laubinger and I (Marcel Quint, speaker), for example, focus on the interaction of proteoforms with nucleic acids to understand how plant populations generate diversity on the level of transcriptional responses to environmental stimuli. Extending SNP2Prot’s within species focus to orthologous proteins between species offers an opportunity to connect CRC 1664 with iDiv to bridge different scales of biodiversity research.
SNP2Prot is supported by MLU and partner institutes (UL, Leibniz Institute of Plant Biochemistry Halle, Leibniz Institute of Plant Genetics and Crop Plant Research in Gatersleben).

